Fig. 2.
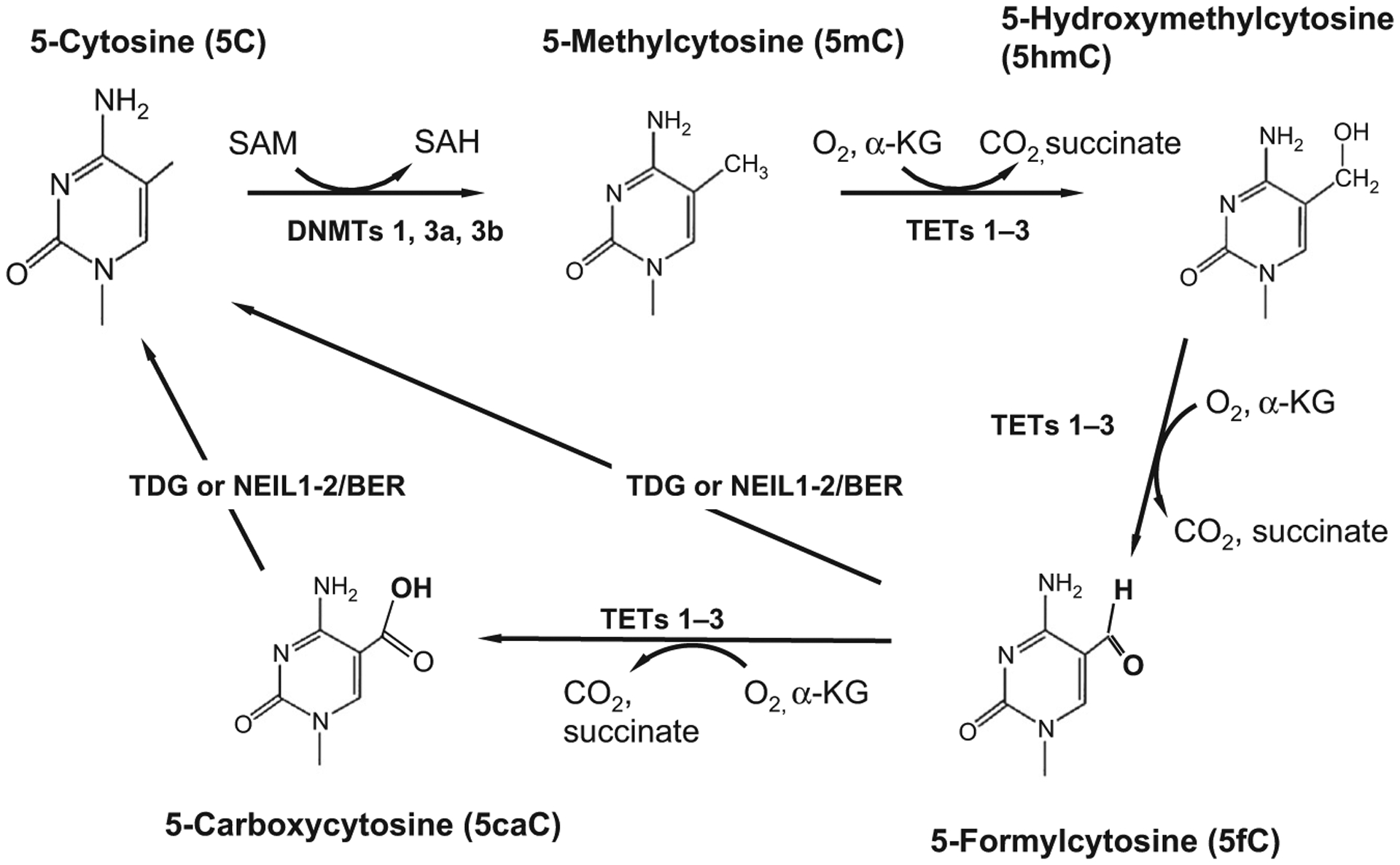
The DNA demethylation pathway. The DNA methyltransferase enzymes (DNMTs 1, 3a, and 3b) that catalyze DNA methylation utilize the universal methyl donor SAM (S-adenosylmethionine) to transfer a methyl group to the 5 position of the heterocyclic aromatic ring of cytosine (5C). Methylcytosine (5mC) is a repressive chromatin mark that negatively correlates with gene expression. Cytosine demethylation, which often reactivates transcription, occurs through several steps, the first of which involves hydroxylation of the methylated cytosine. DNA hydroxymethylation is catalyzed by members of the TET (1–3) family of heme-containing Fe(II)/α-ketoglutarate-dependent dioxygenases. TET proteins further oxidize the hydroxymethyl group to form 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Both 5fC and 5caC can be excised by thymine deglycosylase generating an abasic site which is subsequently replaced by base excision repair (BER) enzymes which regenerate the unmodified cytosine base. Unmodified cytosines (5C) are recognized and bound by proteins such as DNMT1 and TET1 that contain a CXXC motif in their zinc finger DNA-binding domain.9 Methyl-binding domain proteins, like MeCP2 and MBD2, bind to 5mC with high affinity.
