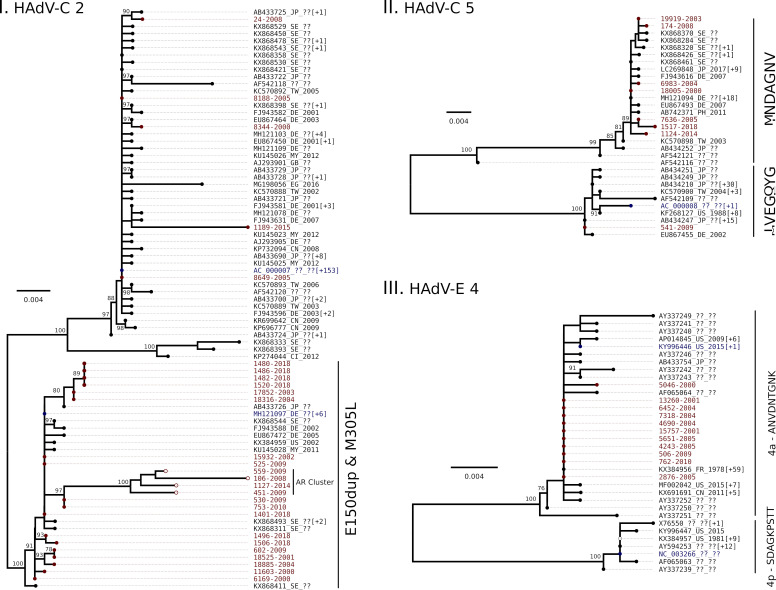
Fig 2. Phylogenetic analyses of HAdV genotypes C2 (I), C5 (II) and E4 (III).
Maximum likelihood phylogenetic trees for sequences from Argentina (red), and from other countries (black). The number between square brackets indicates how many additional HAdV sequences had the same position in the complete tree. The reference sequence(s) for each genotype are in blue. The best fit model was used according to Bayesian Information Criterion (I: HKY+I, II and III: HKY+Γ) for each tree. The numbers near the node represent the branch support (bootstrap % over 1000 pseudoreplica). Amino acid signature or particular features are described at the right of each cluster. I. AR Clade: sequences with an amino acid substitution E to K in the duplicated codon. III. Hollow bullets: sequence with a 4 amino acid deletion at positions 192 to 195.