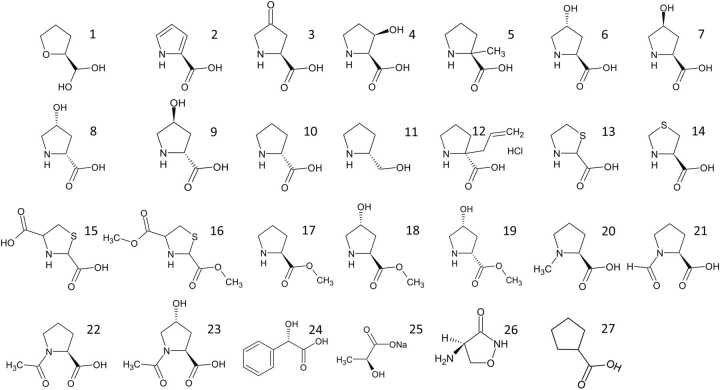
Figure 2.
Structures of the proline analogs screened in crystallo against PYCR1.1, (S)-(−)-tetrahydro-2-furoic acid; 2, pyrrole-2-carboxylic acid; 3, 4-oxo-l-proline, 4, cis-l-3-hydroxyproline; 5, α-methyl-l-proline; 6, trans-4-hydroxy-l-proline; 7, cis-4-hydroxy-l-proline; 8, cis-4-hydroxy-d-proline; 9, trans-4-hydroxy-d-proline; 10, d-proline; 11, R-(−)-2-pyrrolidinemethanol; 12, (S)-α-allyl-proline; 13, thiazolidine-2-carboxylate; 14, l-thiazolidine-4-carboxylate; 15, 1,3-thiazolidine-2,4-dicarboxylate; 16, dimethyl 1,3-thiazolidine-2,4-dicarboxylate; 17, l-proline methyl ester; 18, l-4-hydroxyproline methyl ester; 19, cis-4-hydroxy-d-proline methyl ester; 20, N-methyl l-proline; 21, N-formyl l-proline; 22, N-acetyl l-proline; 23, trans-1-acetyl-4-hydroxyl-l-proline; 24, l-(+)-mandelic acid; 25, sodium-l-lactate; 26, d-cycloserine; 27, cyclopentanecarboxylate.