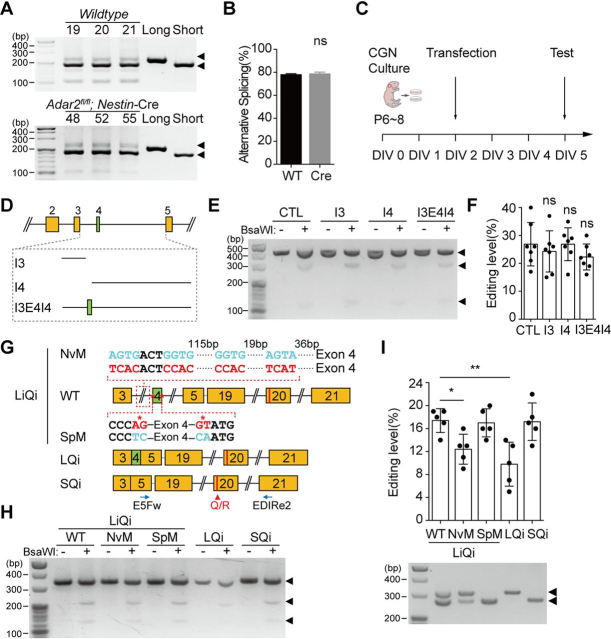
Figure 7.
Alternative splicing of exon 4 regulates Q/R editing at exon 20.A, alternative splicing of exon 4 in brains of WT and Adar2 cKO mice. B, quantification of alternative splicing levels in brains of WT (78.0 ± 1.0%, n = 3) and Adar2 cKO (78.7 ± 1.5%, n = 3) mice. Data are shown as mean ± S.D. ns, not significant; Student's t test, two-tailed. C, schematic demonstration of the experiments conducted in CGNs. D, schematic demonstration of the intronic segments spliced out in alternative splicing. E and F, Q/R editing efficiencies of the editing segment when cotransfected with I3 (24.3 ± 7.4%, n = 7), I4 (26.9 ± 5.9%, n = 7), I3E4I4 (22.3 ± 4.6%, n = 7), and vehicle control (CTL) (26.9 ± 7.8%, n = 7) in CGNs. Data are shown as mean ± S.D. (error bars). ns, not significant; one-way ANOVA, Tukey's post hoc test. G, schematic demonstration of WT and mutant minigene constructs fuzing the editing segment and splicing segment. E5Fw and EDIRe2 are primers for amplification. H and I, Q/R editing of fusion minigenes in CGNs. The Q/R editing efficiency was not changed in SQi (17.2 ± 3.3%, n = 5) and SpM (17.0 ± 2.4%, n = 5) but was suppressed in LQi (9.8 ± 3.8%, n = 5) and NvM (12.4 ± 2.6%, n = 5) mutant, compared with the LiQi control (17.4 ± 2.1%, n = 5). Lower panel in (H) shows the splicing levels corresponding to the upper editing event. Data are shown as mean ± S.D. (error bars). *p < 0.05; **p < 0.01; one-way ANOVA, Tukey's post hoc test.