Figure 2.
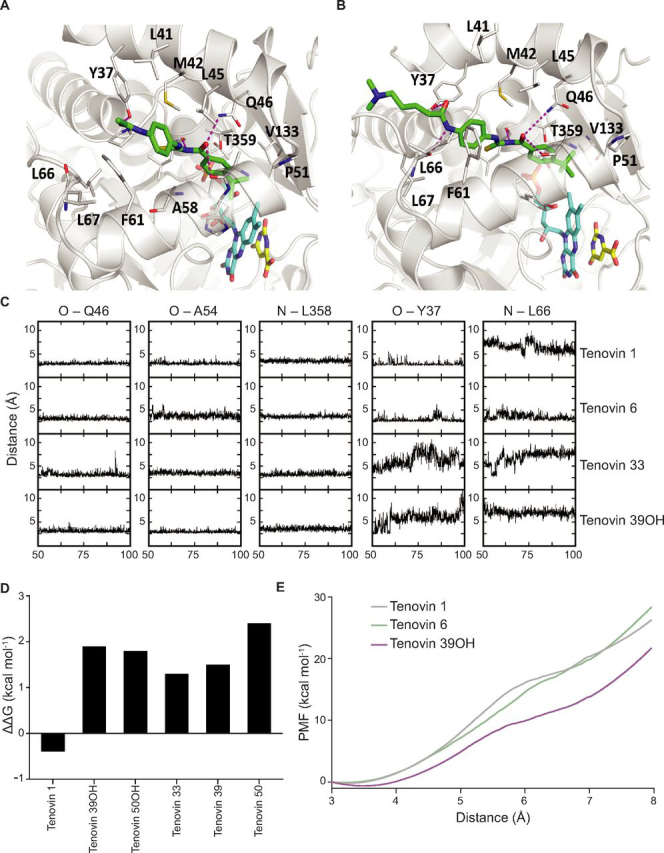
Modeling the interaction between the tenovins and DHODH. Shown is the predicted binding mode of tenovin 1 (A) and tenovin 6 (B) in complex with DHODH. The enzyme DHODH is shown as a cartoon (gray color), and the cofactor FMN (cyan carbon), the substrate DHO (yellow carbon), and bound tenovins (green carbon) are shown as thick lines. The tenovin-binding pocket residues are shown as thin lines, and the hydrogen bonds between DHODH and tenovins 1 and 6 are shown as dashed lines (magenta). C, time evolution of the DHODH–tenovin interactions. Distances between the atom pairs are calculated for the conformations sampled during the second half of the simulations. D, computed free energies differences between the binding of tenovins to DHODH in the membrane relative to the binding of tenovin 6 to DHODH in the membrane (negative value implies tighter binding of a particular tenovin relative to tenovin 6); the FEP/MBAR method was used. E, the free energies required to pull tenovin 1, 6, or 39OH from the binding pocket on DHODH. Positions of the three tenovins relative to DHODH are shown in Fig. S1C.
