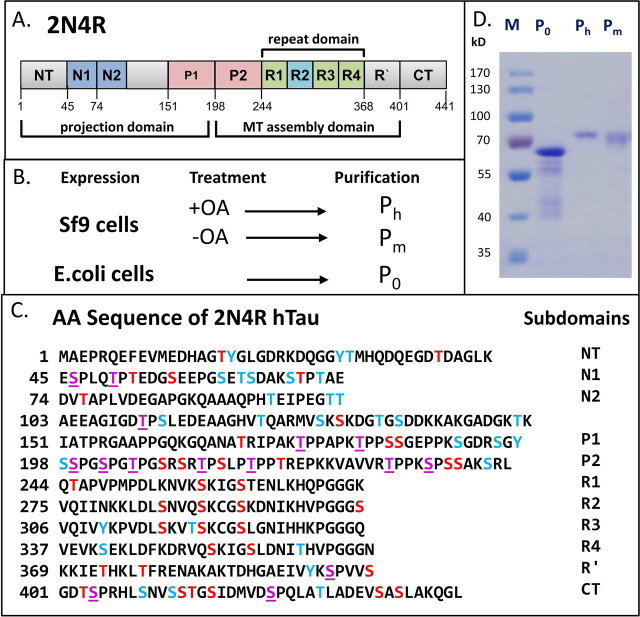
Figure 1.
Overview: Phosphorylation of hTau-2N4R expressed in Sf9 cells and E. coli.A, diagram of domains of hTau40 (2N4R or Tau-F, Uniprot ID P10636-8), the largest human isoform of CNS Tau consisting of 441 residues with the three alternatively spliced inserts N1, N2, and R2. The N-terminal half represents the projection domain, and the C-terminal half contains the four pseudo-repeats (R1–R4) which, in combination with their flanking domains P2 and R′, represent the microtubule assembly domain. B, schematic representation of production of unphosphorylated Tau (P0) in E. coli (prokaryote) and hyperphosphorylated Tau (Pm and Ph) in Sf9 cells (eukaryote). Okadaic acid (OA) treatment increases the phosphorylation of Tau which yields Ph-Tau. C, amino acid sequence of Tau (2N4R, 441 residues) showing phosphorylation sites identified in this study (red and purple letters; purple = Pro directed)) and further potential phosphorylation sites (not detected so far, blue letters) (total of 85 sites, 45 Ser, 35 Thr, and 5 Tyr). Note that only a minority of phosphorylation sites of 37% was detected in the N-terminal part of the sequence (up to residue ∼150, compared with 71% in the C-terminal part of the protein). Note also that none of the five Tyr residues was phosphorylated. D, SDS-PAGE analysis stained with Coomassie Blue showing P0 Tau purified from E. coli and Pm- and Ph-Tau purified from Sf9 cells. Note the upward shift in Mr value with increasing phosphorylation, from 55 kDa (P0-Tau) to 68 kDa for Ph-Tau (compared with the theoretical molecular mass values of ∼45,850 Da and ∼46,863 Da). This shift is characteristic for AD-Tau.