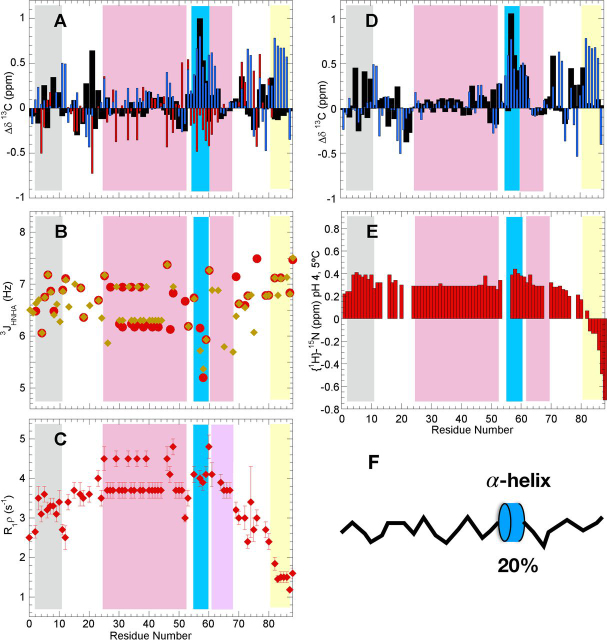
Figure 2.
Orb2A PLD is largely disordered at pH 4, save a short α-helix in residues 55–60. In all panels, the different zones of the Orb2 PLD are shaded as follows: N-terminal hydrophobic stretch, gray; Q/H-rich, cyan; residues 55–60, blue; H·Q-rich, cyan, G-rich, yellow. In the Q/H or H·Q-rich regions, most of the values shown come from averaging of overlapped His or Gln peaks. A, conformational chemical shifts (Δδ) of Orb2 PLD at pH 4, 25°C. 13Cα, black bars; 13CO, blue bars; 13Cβ, red bars. Experimental uncertainties are ≤ ± 0.04, 0.02, and 0.08 ppm for 13Cα, 13CO, and 13Cβ, respectively. B, 3JHNHA coupling constants at pH 4, 25°C. Values of 5 Hz or less are indicative of α-helix; higher values, coil or extended conformations. Values shown in gold were measured by peak height using the Sparky program. Values in red were obtained using the peak integral function of Topspin 4.0. C, transverse relaxation rates in the rotating frame (R1ρ) at pH 4.0, 25°C. D, conformational chemical shifts (Δδ) of Orb2 PLD at pH 4, 5°C. 13Cα, black bars; 13CO, blue bars. The experimental uncertainties are generally ≤ ± 0.05 ppm for both 13Cα and 13CO. E, {1H}-15N NOE at pH 4.0, 5.0°C. Values approaching 0.85 indicate stiffness on fast ps/ns and are typical of well-folded rigid proteins; values less than 0 indicate a high flexibility. Uncertainties are about ± 0.01. F, schematic diagram of Orb2 PLD at pH 4 featuring a flexible and disordered conformational ensemble with a short, modestly populated α-helix spanning residues 55–60.