Figure 1.
Protein composition of broken replication forks
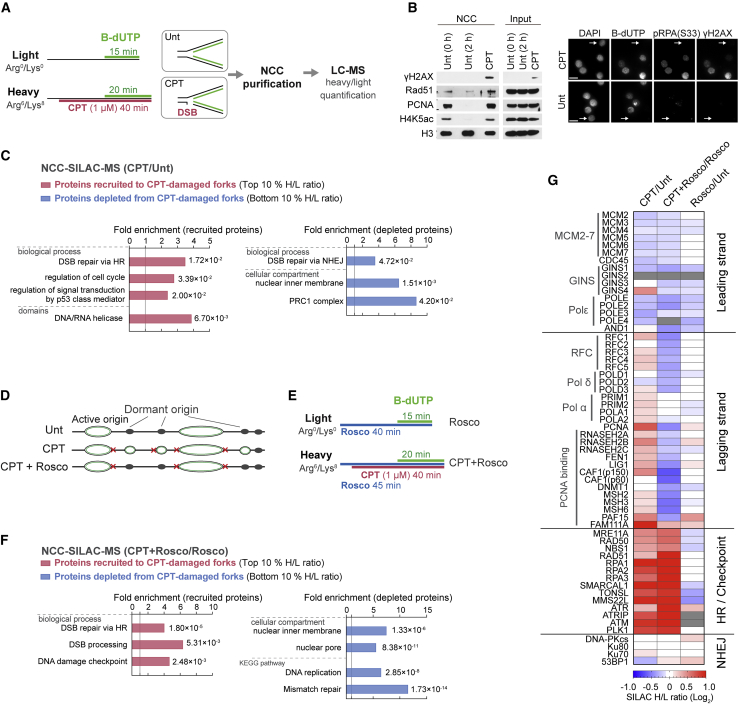
(A) SILAC-NCC-MS strategy for proteomics analysis of broken replication forks. Cells were released from a single thymidine block into mid-S phase and labeled with biotin-dUTP (b-dUTP) in the absence (untreated [Unt]) or presence of CPT (1 μM) prior to NCC purification.
(B) Left: NCC pull-downs analyzed by western blotting. Unt samples were harvested immediately after b-dUTP labeling (0 h) or 2 h. Right: b-dUTP-labeled S-phase cells show CPT-induced DNA damage (γH2AX and pRPAS33). An arrowhead indicates b-dUTP-negative cells (outside of S phase). Scale bar, 15 μm.
(C) GO analysis of proteins recruited (top 10% based on H/L ratio) or depleted (bottom 10% based on H/L ratio) at CPT-damaged forks. All identified proteins were used as background. p vvalue is shown with fold enrichment.
(D) Illustration of dormant origin firing in response to CPT and suppression of new origin firing by roscovitine (rosco).
(E) Experimental design for NCC-SILAC-MS using roscovitine to block CPT-induced dormant origin firing.
(F) GO analysis as in (C).
(G) Heatmap of replication and DDR factor enrichment, indicating the mean of three independent experiments. Not detected is indicated by gray coloring.
See also Figure S1.