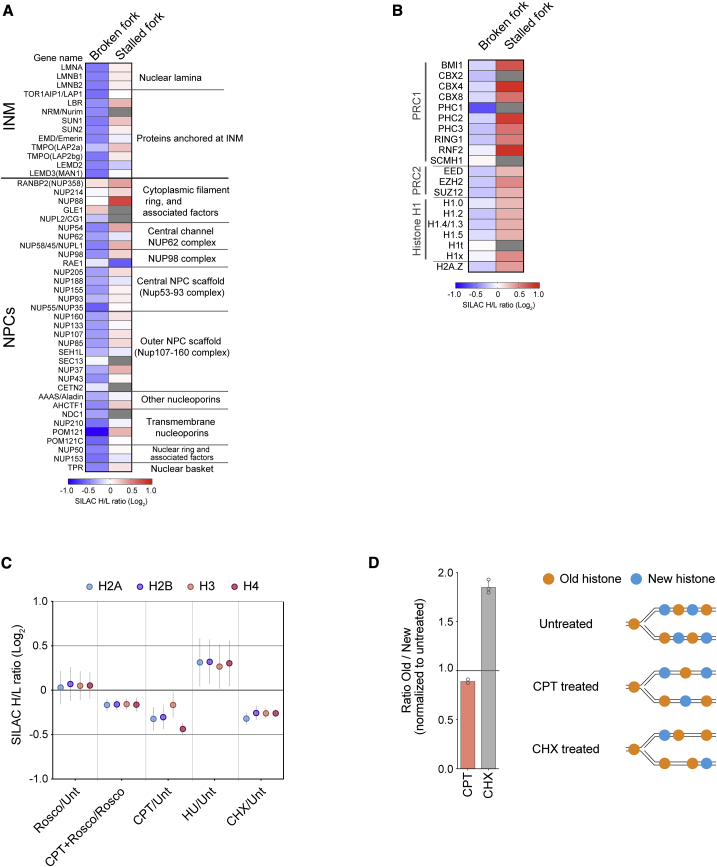
Figure 3.
Distinct chromatin environment at broken and stalled replication forks
(A and B) Heatmap showing enrichment of inner nuclear membrane (INM) proteins (A), nuclear pore complex (NPC; A), and chromatin regulators (B) at broken (CPT+Rosco/Rosco) and stalled (HU/Unt) replication forks. The mean of three independent experiments is shown.
(C) Enrichment of canonical histones across all NCC-SILAC-MS data (see Figures S7A and S7B for details). Mean is shown with SEM; n = 3. CHX, cycloheximide.
(D) Left: the ratio of newly synthesized (new) and old recycled (old) histone H4 at replication forks isolated from CPT- and CHX-treated cells. New and old histones were analyzed by pulse-SILAC labeling followed by NCC-MS (Figure S3A). The CHX dataset is from Alabert et al. (2015). Mean is shown with SEM; n = 3 (CHX), 2 (CPT). Right: illustration of new and old histones in nascent chromatin.
See also Figure S3.