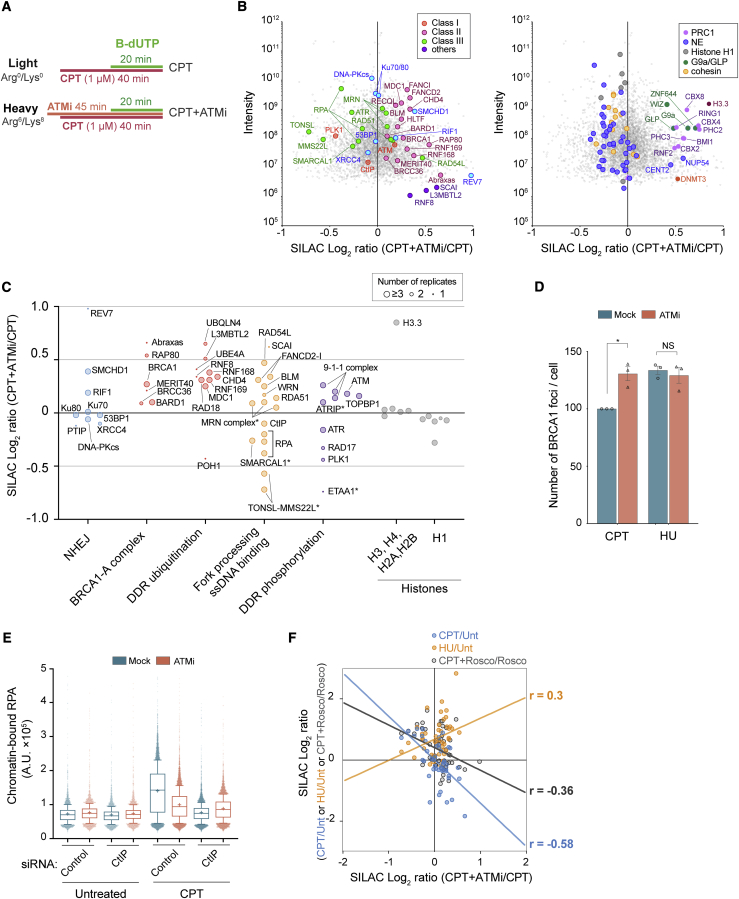
Figure 4.
ATM inhibition rewires the broken replication fork proteome toward a stalled fork response
(A) Experimental design of NCC-SILAC-MS analysis of ATM function at CPT-damaged forks (CPT+ATMi/CPT). The ATMi AZD0156 (250 nM) was added 5 min before CPT (1 μM) treatment.
(B) Enrichment of DDR (left) and chromatin (right) proteins, shown as log2 SILAC ratios of CPT+ATMi (heavy) over CPT (light) (CPT+ATMi/CPT). The mean of six independent experiments is shown. NE, nuclear envelope.
(C) Enrichments of DDR proteins from (B), grouped according to function. Symbol size indicates the number of replicates in which a protein was identified. ∗, RPA binding proteins.
(D and E) High-content microscopy of U-2-OS cells exposed to CPT or HU for 1 h with or without ATMi added 5 min before. Pre-extracted cells were stained for γH2AX and BRCA1 (D) or RPA (E).
(D) BRCA1 foci in γH2AX positive cells are shown relative to cells treated with CPT alone. Error bars indicate SEM; n = 3. Individual measurements are indicated by dots and correspond to the mean of more than 1,092 cells. ∗p = 0.0303; NS, not significant by ratio-paired two-sided t test. See the gating strategy for γH2AX positive cells in Figure S4I.
(E) RPA intensity shown as mean (+), with whiskers indicating 10th–90th percentiles; from left, n = 3,858, 3,402, 3,398, 3,249, 8,946, 9,509, 8,525, and 8,068 cells. See the gating strategy for RPA-positive cells in Figure S4J.
(F) Correlation plot showing DDR proteins identified in NCC-SILAC-MS as indicated. Pearson correlations (r) are shown.
See also Figure S4.