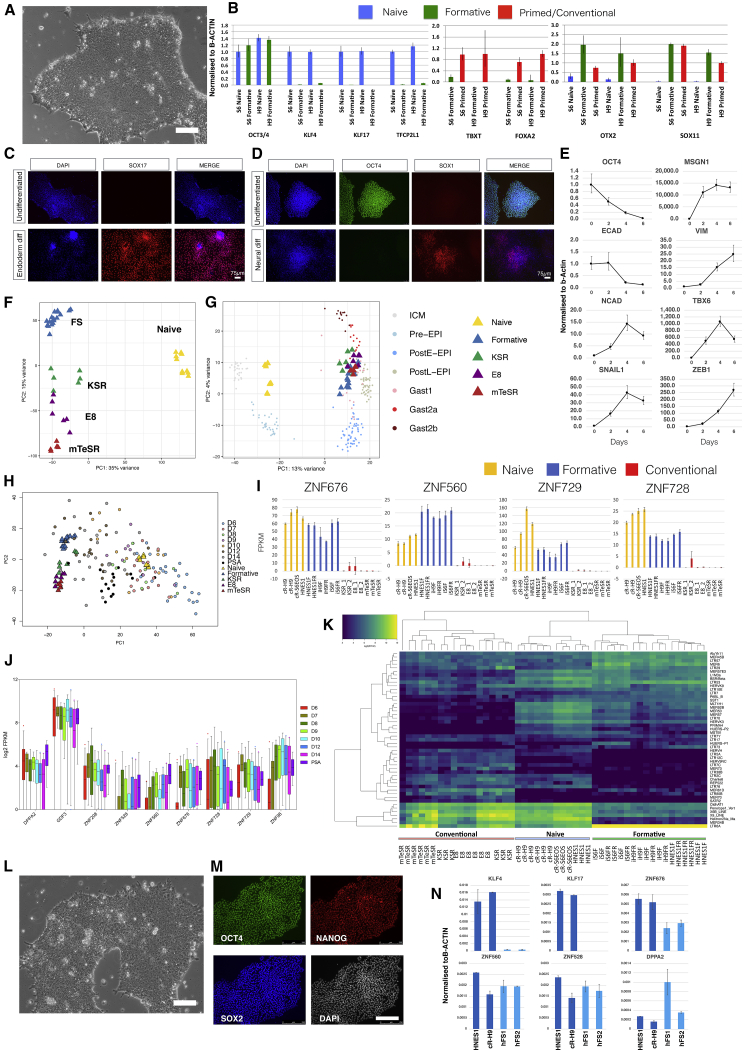
Figure 7.
hFS-like Cells Established from Naive ESCs and Embryos
(A) Morphology of human AloXR cells derived from naive hPSCs. Scale bar, 100 μm.
(B) qRT-PCR expression analysis of marker genes in two human FS (hFS) cell lines compared with naive and conventional (primed) hPSCs. Error bars represent SD from technical triplicates.
(C) SOX17 immunostaining of hFS cells after endoderm induction.
(D) SOX1 immunostaining of hFS cells after neural induction.
(E) qRT-PCR analysis of hFS cells differentiated into paraxial mesoderm for 6 days. Error bars represent SD from technical triplicates.
(F) PCA of hFS cells with naive and conventional hPSCs computed with 11,051 genes identified by median Log2 expression of >0.5.
(G) Projection of hFS cell and conventional PSC samples onto PCA of Macaca ICM/epiblast stages computed with 9,432 orthologous expressed genes.
(H) PCA for cell line populations computed using 922 variable genes across epiblast samples from human embryo extended culture (Xiang et al., 2019) with projection of embryo single cells.
(I) Fragments per kilobase of exon model per million reads mapped (FPKM) values for naive-formative specific genes in naive, formative, or conventional hPSCs.
(J) Boxplots of naive-formative specific gene expression in human epiblast stages and primitive streak anlage (PSA).
(K) Heatmap of differentially expressed transposable elements between naive, formative, and conventional samples.
(L) Morphology of FS cells derived directly from human embryo. Scale bar, 100 μm.
(M) Immunostaining of OCT4, SOX2, and NANOG in embryo-derived hFS cells. Scale bar, 250 μm.
(N) qRT-PCR analysis of embryo-derived hFS cells. Error bars represent SD from technical duplicates.