Figure 1.
Differentiation of hypertrophic chondrocytes from iPSCs
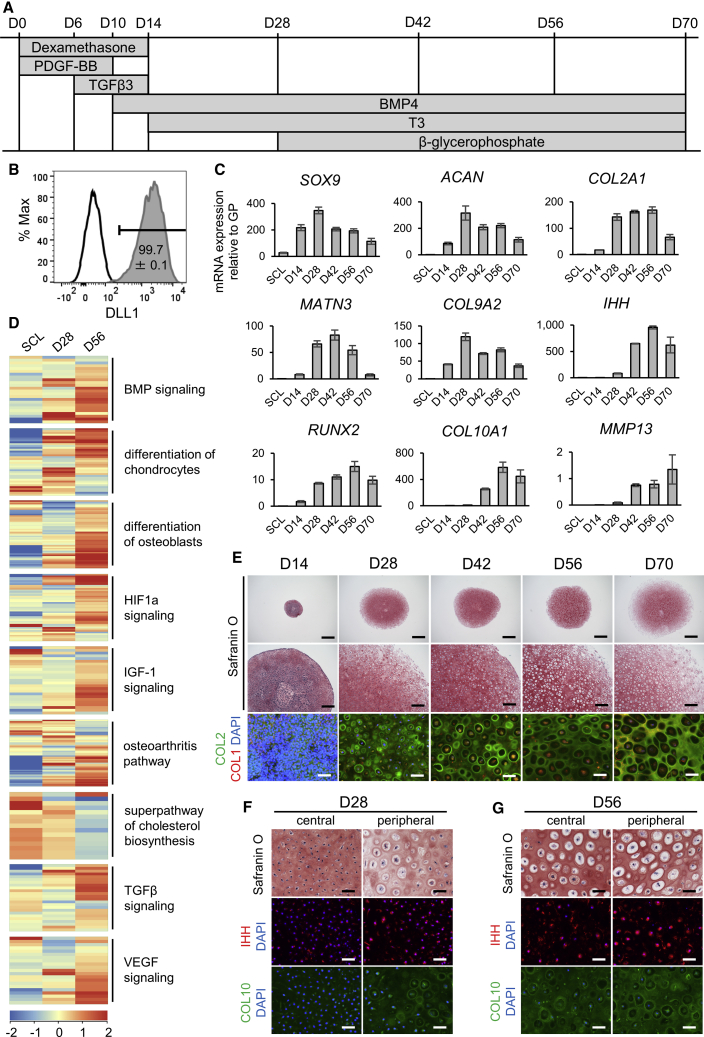
(A) Protocol of hypertrophic chondrocyte induction (HI) in 3D pellet culture from sclerotome (SCL) cells (D0).
(B) Representative result of DLL1-positive cells (compared with isotype control) on day 2 of sclerotome induction (SI), with the mean and SEM (standard error of the mean) of n = 4 independent experiments displayed.
(C) mRNA expression of chondrocyte markers of different stages over time from SCL on day −1 to day 70 of HI by qPCR (top row, early markers; middle row, proliferating to prehypertrophic markers; bottom row, hypertrophic markers). Values are shown as mean ± SEM (n = 4 independent experiments), relative to the mean of six pieces of a human distal femoral growth plate (GP).
(D) Heatmap of averaged normalized intensity values of gene expression in chondrocyte hypertrophy-related pathways from IPA in SCL, day-28, and day-56 samples (n = 3 independent experiments).
(E) Safranin O staining (top and middle rows) and COL2 (green) with COL1 (red) immunostaining (bottom row) of pellets from days 14–70 of HI. Scale bars, 1 mm (top row), 200 μm (middle row), 50 μm (bottom row).
(F and G) Safranin O and IHH or COL10 immunostaining on day 28 (F) and day 56 (G) of HI in the central or peripheral area of each pellet. Scale bars, 50 μm.
All results shown are from experiments using the 414C2 wild-type iPSC line. For (E), (F), and (G), similar results were obtained in n = 3 independent experiments. See also Figure S1 and Table S1.