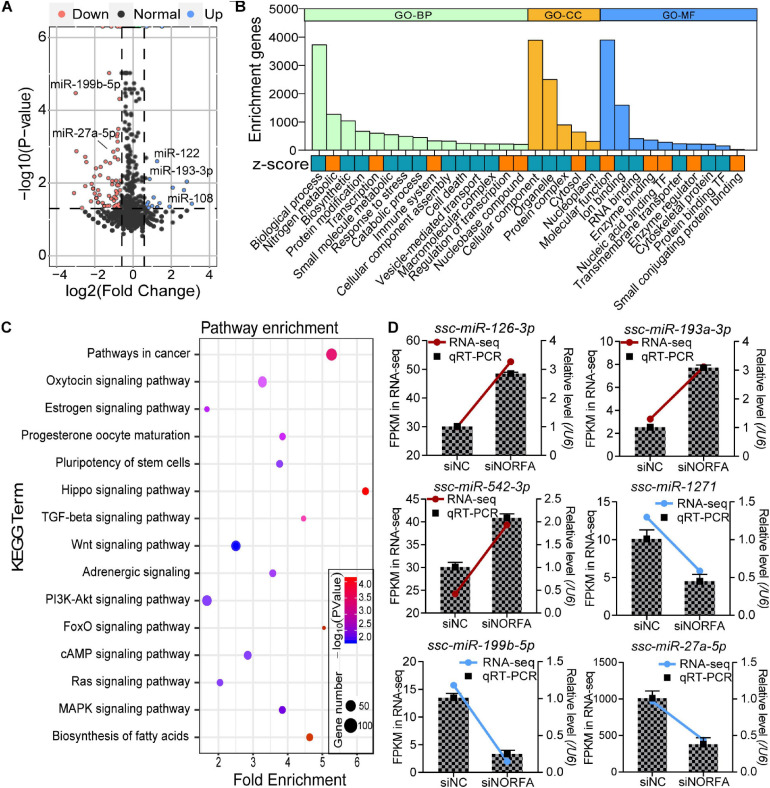
FIGURE 5.
Identification and functional annotation of DEmiRNAs in porcine GCs treated with NORFA-siRNA. (A) Volcano plot showing the expression profile of DEmiRNAs in NORFA-reduced porcine GCs with the following criteria FDR < 0.05 and |Log2(fold change)| ≥ 0.59. Up- and down-regulated genes are labeled in blue and pink; the black points indicate normal genes. Several important DEmiRNAs were also indicated in the volcano plot. (B) Gene Ontology analyses of DEmiRNAs in porcine GCs treated with NORFA-siRNA. The columns in green, yellow, and blue indicate the enriched terms of BP (biological process), CC (cellular components), and MF (molecular function) categories. The significant GO terms with their full names were listed below and their expression patterns were estimated by z score, which were labeled in orange (up-regulated) and blue (down-regulated). (C) KEGG pathway enrichment analyses of DEmiRNAs in NORFA-inhibited porcine GCs and significant KEGG terms were also presented. (D) Six DEmiRNAs (three up- and three down-regulated) that are crucial for ovarian GCs were selected, and their expression patterns in porcine GCs were validated by qRT-PCR. The color lines depict the expression patterns of DEmiRNAs determined by RNA-seq, which refers to the left y-axis (red and blue indicate up- and down-regulated). The bars represent the results of qRT-PCR by referring to the right y-axis (mean ± S.E.M.; n = 3).