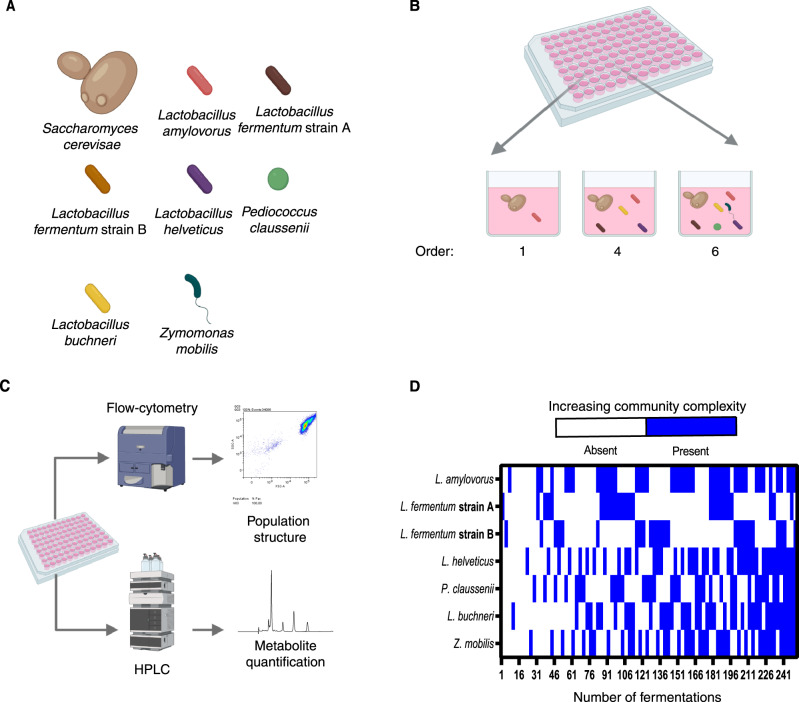
Fig. 1. Schematics of the combinatorial assembly experimental setup.
A Microbial species used in this study. These species make up >80% of the diversity found in fuel ethanol fermentations in relative abundance. B Initially, pairwise fermentations between yeast inoculum and all available bacteria were performed. This results in a combinatorial assembly of order 1. In parallel, every pairwise assembly was also mixed with every other bacterial monoculture, resulting in a set of 51 different fermentations or order 2. This was performed for every possible combination of bacterial species and yeast, resulting in 258 fermentations, and a total order of 6 (i.e., a community containing up to six different bacterial species and yeast). The ratio between all bacterial species was kept at 1 for every cultivation, and the yeast:bacteria ratio was 100:116. C After fermentation was completed, the community structure was analysed via flow cytometry, and the fermentation metabolites were quantified via HPLC. D An overview of the microbial compositions generated via the combinatorial assembly throughout the 258 fermentations. This figure was created by FSOL using Biorender (https://app.biorender.com/).