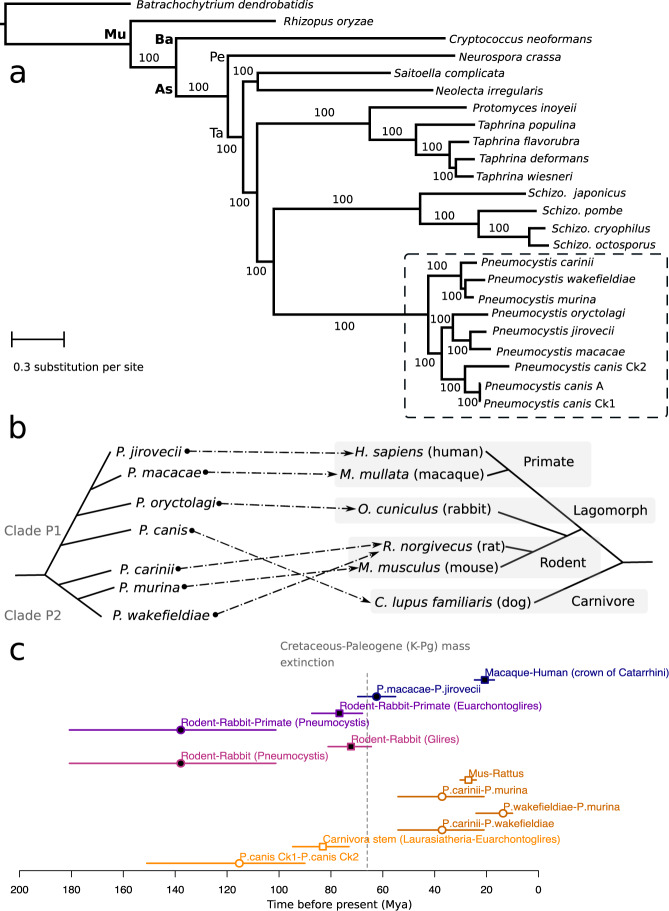
Fig. 2. Phylogeny and divergence times of Pneumocystis species.
a Maximum likelihood phylogeny constructed using 106 single-copy genes based on 1000 replicates from 24 annotated fungal genome assemblies including nine from Pneumocystis (highlighted with a dashed box). Only one assembly is shown for each species except there are three for P. canis (assemblies Ck1, Ck2, and A). Bootstrap support (%) is presented on the branches. The fungal major phylogenetic phyla and subphyla are represented by their initials: As (Ascomycota), Ba (Basidiomycota), Pe (Pezizomycotina), Mu (Mucoromycota), and Ta (Taphrinomycotina). b Schematic representation of species phylogeny and association between Pneumocystis species and their respective mammalian hosts. The dashed arrows represent the specific parasite-host relationships. c Divergence times of Pneumocystis species and mammals (n = 12 taxonomic clades analyzed). Divergence time medians are represented as squares for hosts and as circles for Pneumocystis, and the horizontal lines represent the 95% confidence interval (CI) error bars, which are color-coded the same for each Pneumocystis and its host. Closed elements represent nodes that are different in term of divergence times (nonoverlapping confidence intervals) whereas open elements represent nodes with overlapping confidence intervals. Catarrhini, taxonomic category (parvorder) including humans, great apes, gibbons, and Old-World monkeys. Euarchontoglires, superorder of mammals including rodents, lagomorphs, treeshrews, colugos, and primates. Glires, taxonomic clade consisting of rodents and lagomorphs. Laurasiatheria, taxonomic clade of placental mammals that includes shrews, whales, bats, and carnivorans. Mya, million years ago. K-Pg, Cretaceous-Paleogene. The dotted vertical line representing the K-Pg mass extinction event at 66 mya is included for context only.