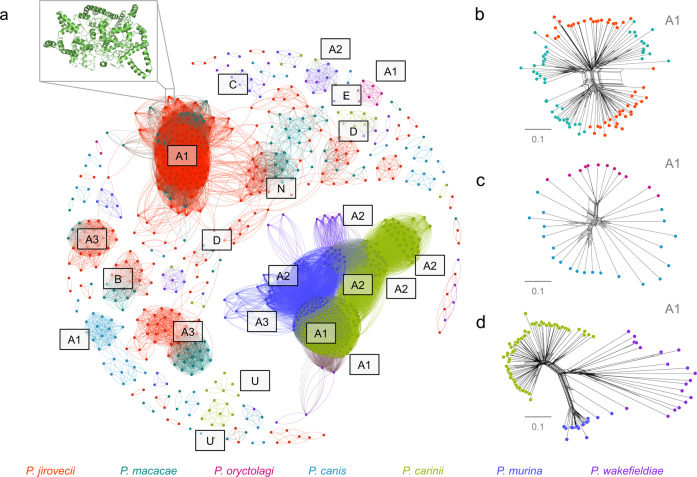
Fig. 4. Clustering of Pneumocystis major surface glycoproteins (Msg).
a Graphical representation of similarity between 482 Msg proteins from seven Pneumocystis species generated using the Fruchterman Reingold algorithm. A 3-D model of a representative member of Msg-A1 protein family (NCBI locus tag T551_00910) generated using DESTINI is presented in the upper left insert. Individual protein sequences are shown as dots and color-coded by species as shown at the bottom of the figure. The edge between two dots indicates a global pairwise identity equal or greater than 45%. The letters represent Msg families (A to E) and subfamilies (A1 to A3). N and U letters represent potentially novel Msg sequences (relative to our prior study35) and unclassified sequences, respectively. For sake of clarity only the major clusters were annotated. b Phylogenetic network of a subset of Msg-A1 family (n = 97) in primate Pneumocystis including P. jirovecii (red) and P. macacae (dark cyan) suggesting recombination events at the root of the network. Nodes with more than two parents represent reticulate events. Bars represent the number of amino acid substitutions per site. c Phylogenetic network of Msg family A1 (n = 33) in P. oryctolagi (red violet) and P. canis (light blue). d Phylogenetic network of Msg-A1 family (n = 113) in rodent Pneumocystis including P. carinii (green), P. murina (dark blue), and P. wakefieldiae (blue violet). The network data are available at 10.5281/zenodo.4450766.