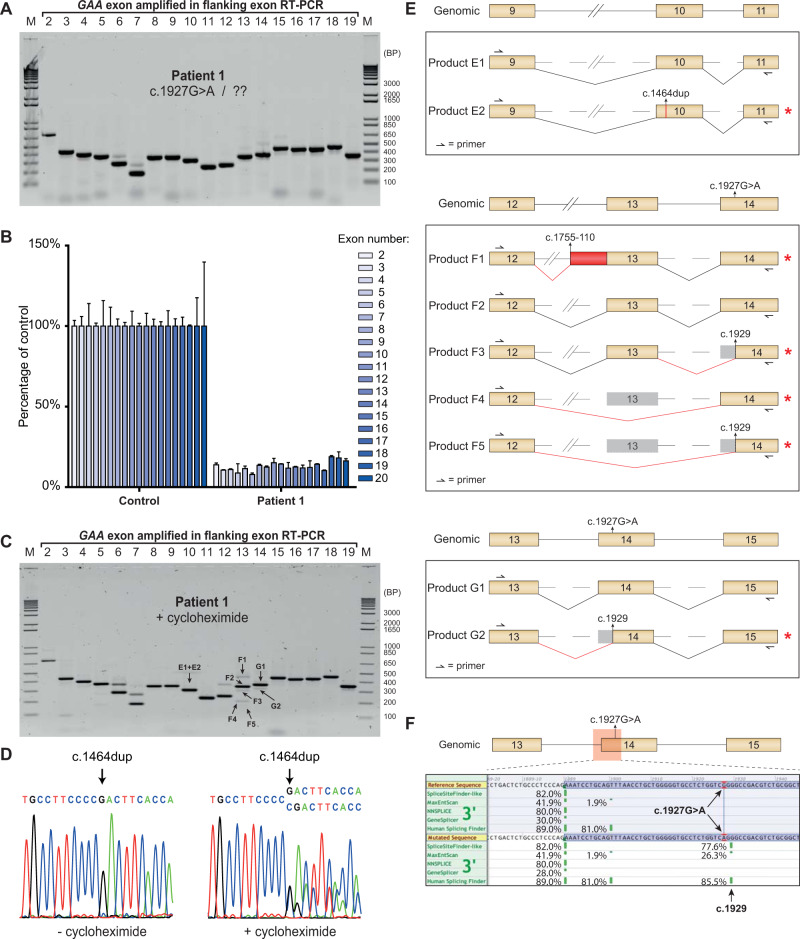
Fig. 2. Analysis of aberrant splicing products in Patient 1.
a Flanking exon RT-PCR of Patient 1 primary fibroblasts. b Exon-internal RT-qPCR for all coding exons of Patient 1 compared to healthy control. Data are shown as mean ± S.D. from three technical replicates. c Same as in a, but after cycloheximide treatment. d Sequencing results of flanking exon RT-PCR exon 10 sample with and without cycloheximide treatment. e Cartoons of products present in PCR samples for exons 10, 13 and 14. Product numbers refer to product highlighted in c. Red asterisks indicate products that undergo NMD. Red boxes indicate non-canonical sequences present in the GAA mRNA transcripts. Gray area’s highlight skipping of canonical GAA mRNA sequences. Primer targets are indicated. f Output of splice prediction performed in Alamut®. Five splice site prediction algorithms are shown for the area indicated in red, with or without the presence of the c.1927G>A variant. The location of the cryptic donor site utilized in products F3, F5 and G2 is highlighted.