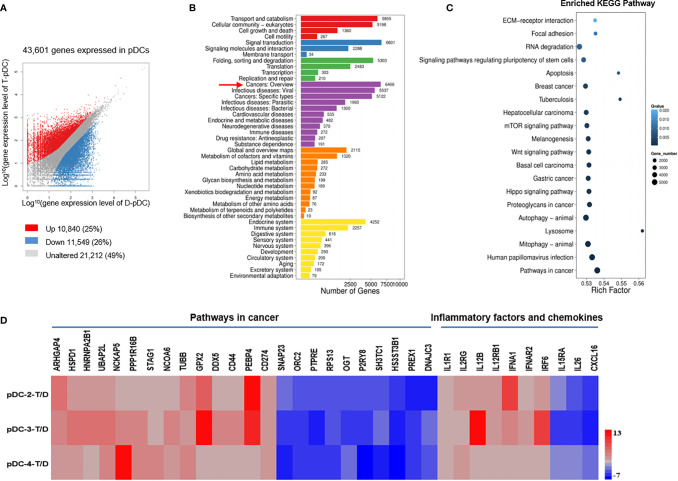
Figure 4.
DEGs in pDCs are associated with tumor development. Analysis of gene expression in tumor and distal pDCs and identification of significant pathways regulated by the DEGs. (A) Scatter plot of DEGs: X and Y axes represent log10-transformed gene expression levels. Upregulated, downregulated, and unchanged genes are presented in red, blue, and gray, respectively. (B) Pathways regulated by the DEGs: The X-axis represents the number of DEGs, whereas the Y-axis indicates the functional classification. (C) Pathway functional enrichment of DEGs: The X-axis represents the enrichment factor and the Y-axis signifies the pathway name. The color indicates the q-value (high: white; low: blue). A lower q-value represents a more significant enrichment. Point size indicates DEG number. Rich Factor refers to the value of the enrichment factor, which is the quotient of the foreground value (the number of DEGs) and the background value (total number of genes). The larger the value, the more significant the enrichment. (D) RNA-Seq analysis of DEGs in tumor versus distal pDCs. Data from three separate tumor pDCs (T, experimental group) versus data from three separate distal pDCs (D, control group). Many of the DEGs in pDCs were associated with tumor development and metastases.