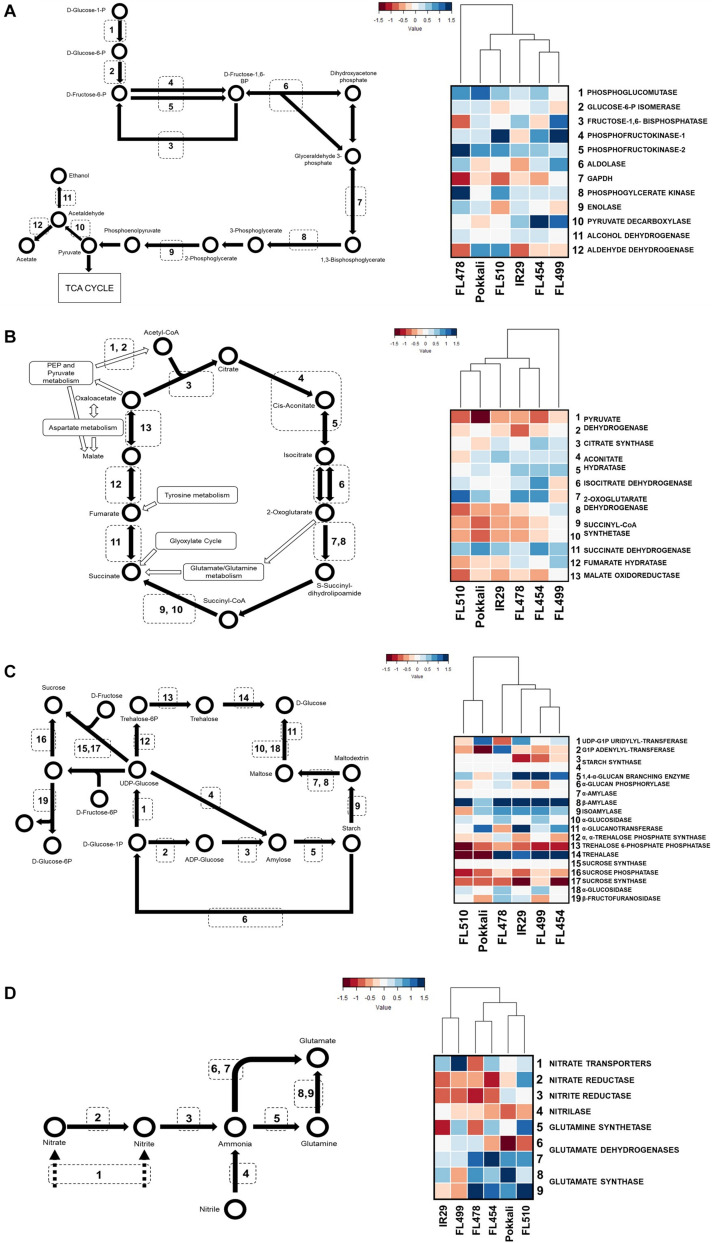
FIGURE 8.
Metabolic similarities based on transcriptome profiles. KEGG models of metabolic pathways are shown on the left panel for glycolysis (A), TCA cycle (B), starch/sucrose metabolism (C), and nitrogen assimilation (D). Transcripts for enzymes in each step were mapped against each pathway. Pathway activities and the pattern of similarities across genotypes are depicted by hierarchical clustering dendrograms based on maximum log2-fold change in transcript abundances at the time-point with the widest difference from control. FL510 and Pokkali share similarities in repression of the carbon metabolism pathways, while nitrogen assimilation is induced in both genotypes. In comparison, the poor genotypes cluster together.