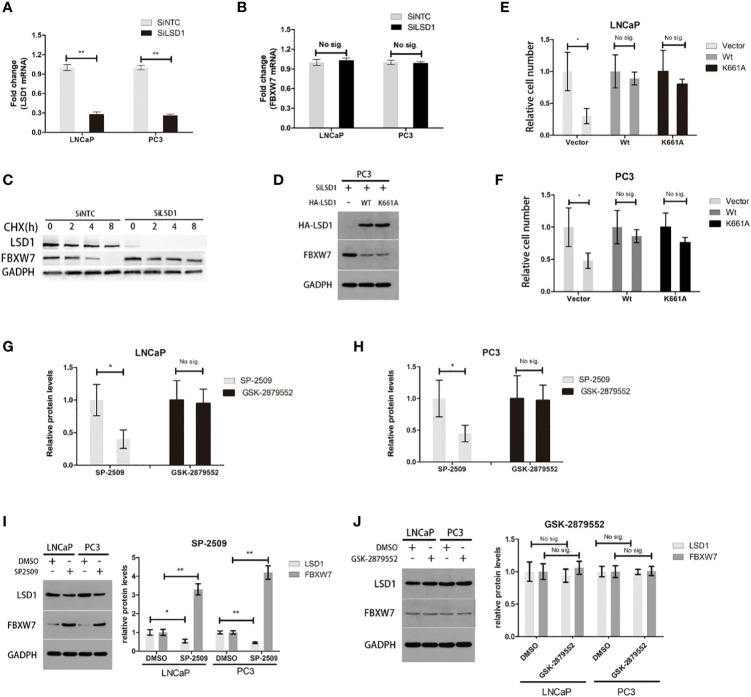
Figure 4.
It is independent of demethylation-function that LSD1 can destabilize FBXW7 in PCa. (A, B) The change of LSD1 and FBXW7 mRNA levels were detected by qPCR after LSD1 knockdown. (C) LSD1 and FBXXW7 half-life were detected by western blotting analysis at various time points, such as 0, 2, 4, and 8h after treatment of CHX. (D) Western blot analysis was used to detect protein expression level of LSD1 and FBXW7 in PC3 cells that were transfected with indicated exogenous protein. (E, F) Relative cell number of LNCaP and PC3 cells after transfected with exogenous wild-type LSD1 or catalytically deficient mutant K661A. Wild-type and catalytically deficient mutant LSD1 both can decrease FBXW7 protein level after transfected with SiLSD1 in PC3 cells. (G, H) Relative cell number of both cell lines after treated with SP-2509 or GSK-2879552 for 72h. (I, J) The relative protein expression level of LSD1 and FBXW7 in both cell lines after the treatment of inhibitors. The simple catalytic inhibitor GSK-2879552 cause no significant change. And SP-2509 cause the LSD1 depletion which results in up-regulation of FBXW7 expression level. This result is absent in GSK-2879552 treated group. Values are expressed as the mean ± SEM.*P < 0.05, **P<0.01, relative to the vector or DMSO group, n = 3. Protein levels were normalized by GADPH.