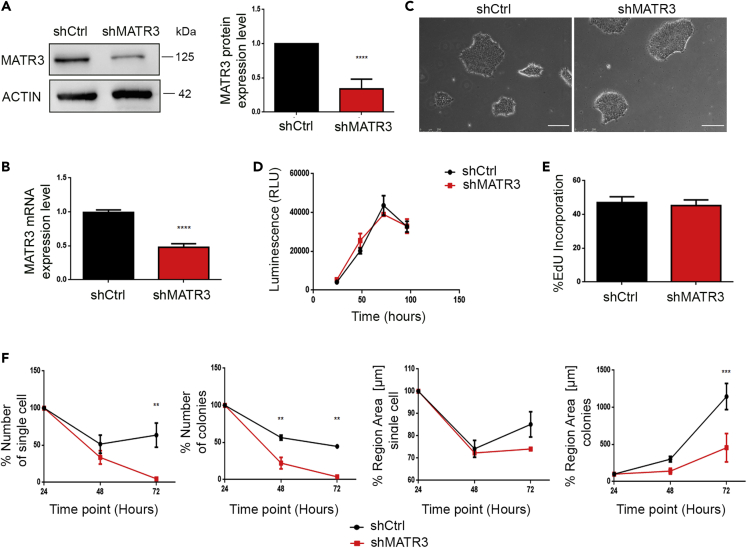
Figure 1.
MATR3 is essential for single-cell growth of hiPSCs
(A) MATR3 protein expression levels were analyzed by western blot (WB) in hiPSCs stably infected with shMATR3 and shCtrl lentiparticles. β-ACTIN was used as housekeeping (left panel). MATR3 levels were quantified by densitometry analysis. The bar plots show the mean values of three independent experiments (right panel). Graph represents a mean of three biological replicates ± SEM. p value was calculated by t test (∗∗∗∗p < 0.0001).
(B) MATR3 RNA expression levels were measured by real-time qPCR in hiPSC line stably infected with shMATR3 and shCtrl lentiparticles. Data are presented as mean of three biological replicates ± SEM. p value was calculated by t test (∗∗∗∗p < 0.0001).
(C) Pictures of shCtrl and shMATR3 hiPSCs (scale bar, 250 μm).
(D) Growth curve of shCtrl and shMATR3 hiPSCs were performed using OZBlue assay. Data are presented as the mean ± SEM of three biological replicates; two-way ANOVA (not significant, p>0.05).
(E) EdU proliferation assay for shMATR3 and shCtrl hiPSCs. Percentage of EdU-incorporating cells was measured by flow cytometry. Data are presented as means ± SEM (n = 2 biological replicates, 6 technical replicates); t test not significant, p>0.05 .
(F) Analyses of shMATR3 and shCtrl hiPSCs following single-cell seeding. Cells were incubated with AP Live Stain at each time point (time point, 24, 48, and 72 h). Images were acquired with the Operetta High Content Screening System using 2× objective 0.08NA. The region areas [μm] for the single cells and for the colonies are reported for each time point. The comparison is between shCtrl and shMATR3 cells. Two-way ANOVA (∗∗ p<0.01; ∗∗∗ p< 0.001).