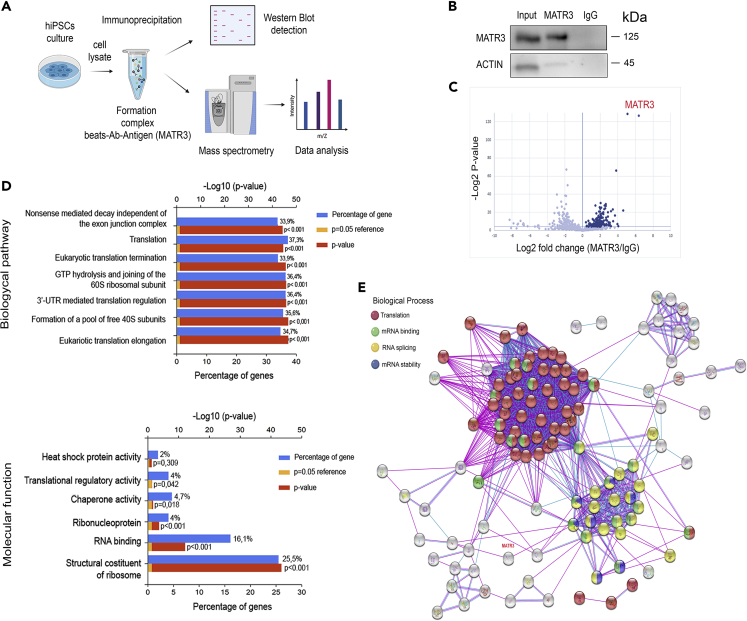
Figure 4.
MATR3 is associated to the translational machinery and RNA processing
(A) Schematic representation of the strategy used for the identification of MATR3 interactors. (B) WB performed on parental hiPSC cell lysates co-immunoprecipitated with MATR3 antibody (ab) and IgG as control. Inputs are also shown (Input). The β-ACTIN signal is present only in the input sample confirming the specificity of MATR3 antibody.
(C) Volcano plot representation of MS analysis results. The ScatterPlot showed 151 proteins significantly enriched in MATR3 (dark blue dots, right of the graph) versus IgG pulldown (left part of the graph), with a t test p value < 0.05 (or > 4.32 when −log2 transformed).
(D) Functional enrichment analysis of MATR3 interactors conducted by FunRich software. Two different GO biological processes analyses, Biological pathway and Molecular function, were used. Significance of the interactions is represented by a red histogram, p < 0.001 (orange with minimum p < 0.05), and the percentage of overrepresentation of the genes is represented by a blue bar.
(E) Protein-protein interactions calculated by STRING interaction database. Only high confidence interactions (interaction score >0.7), as determined by the STRING database, were accepted. Each node represents a protein, and edges represent protein-protein associations. In the graph, proteins involved in translation (red plot), mRNA processing (green plot), RNA splicing (yellow plot), and mRNA stabilization (blue plot) are reported.