Figure 5.
MATR3 regulates the translation efficiency of OCT4, LIN28A, and NANOG mRNAs
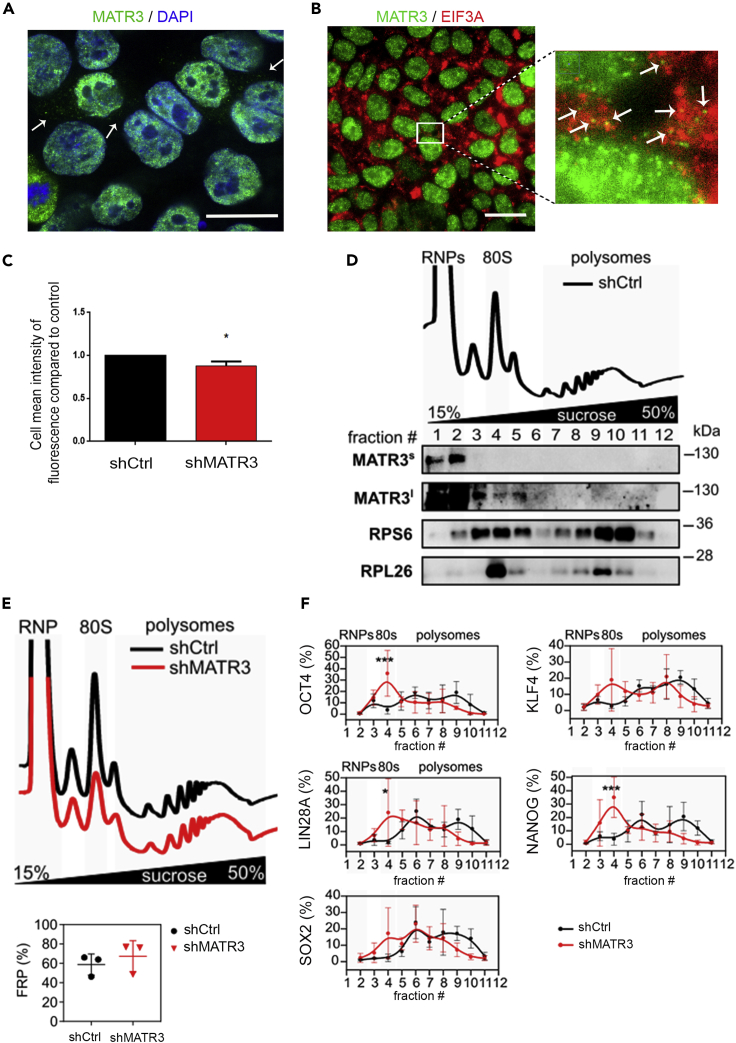
(A) Immunofluorescence pictures to detect the subcellular localization of MATR3 in hiPSCs. Scale bar, 20 μm.
(B) Pictures of hiPSCs stained for MATR3 (green signal) and eIF3a (red signal). Nuclei were stained with DAPI (blue signal). Scale bar, 20 μm. Zoom highlights an overlight between MATR3-EIF3A (orange spots).
(C) Evaluation of de novo protein synthesis measured by O-propargyl-puromycin (OPP) Alexa Fluor 488 incorporations. Quantification performed by Operetta High Content Screening System. Data presented as mean ± SEM of three biological replicates. shMATR3 values are compared with shCtrl (reported as value 1). The statistical significance was calculated by t test, 95% of confidence.
(D) Polysome profiling performed on shCtrl hiPSC and co-sedimentation profiles of MATR3 and ribosome markers RPS6 and RPL26. The signal of MATR3 along the profile is shown for short (MATR3s) and long (MATR3l) exposure times of acquisition. A single biological replicate is shown.
(E) Polysomal profiles of shCtrl and shMATR3 hiPSC and comparison between FRP in shCtrl and shMATR3 (shCtrl: n = 3, shMATR3: n = 3). No significant changes identified with paired two-tailed t test (p = 0.1075).
(F) qRT-PCR analysis for SOX2, OCT4, LIN28A, NANOG, and KLF4 performed on mRNA from fractions obtained from the polysome profiling. Statistical analysis was performed on the mean of three biological replicates, two-way ANOVA (∗p < 0.05; ∗∗∗p < 0.001).