Graphical abstract

Keywords: Computer-aided design, Site-specific protein binder, LRR protein, HER2
Abstract
A protein binder with a desired epitope and binding affinity is critical to the development of therapeutic agents. Here we present computationally-guided design and affinity improvement of a protein binder recognizing a specific site on domain IV of human epidermal growth factor receptor 2 (HER2). As a model, a protein scaffold composed of Leucine-rich repeat (LRR) modules was used. We designed protein binders which appear to bind a target site on domain IV using a computational method. Top 10 designs were expressed and tested with binding assays, and a lead with a low micro-molar binding affinity was selected. Binding affinity of the selected lead was further increased by two-orders of magnitude through mutual feedback between computational and experimental methods. The utility and potential of our approach was demonstrated by determining the binding interface of the developed protein binder through its crystal structure in complex with the HER2 domain IV.
1. Introduction
A protein binder targeting a specific epitope and a binding affinity is crucial to its development as a therapeutic agent since its efficacy is largely affected by a region where it binds on a target and its binding affinity [1], [2]. Experimental approaches comprising repeated rounds of a library construction and screening have been most widely used, but they are labor-intensive and time-consuming, and almost impossible to specify the binding region [3], [4]. The difficulty dramatically emerges if a target is a multi-domain protein with a large size. Especially, in the case of a library-based approach, selection of an initial binder usually determines the fate of a whole process including the in vitro and in vivo experiments.
Computational methods have recently attracted a considerable attention as a promising paradigm to design a protein binder with desired activity. Advances in computing power and algorithms have enabled the prediction of precise energy landscapes, leading to notable successes in computational protein designs [5], [6], [7], [8], [9], [10]. Despite many advances, however, purely computational design of a protein binder with a desired epitope and binding affinity remains a challenge. It has been known that current scoring functions may not be precise enough mainly due to limitations to accurately define the binding free energy landscapes [11]. Furthermore, if a target protein is composed of multi-domains and structurally flexible loops, it is extremely difficult to computationally design a protein binder with a desired epitope and a high affinity. Overall, design of such protein binder by computational method has been limited so far to target proteins with certain “ideal” features such as high secondary-structure content [12].
Human epidermal growth factor receptor 2 (HER2) is a well-known drug target for various cancers, representing a typical multi-domain membrane protein mainly composed of a number of flexible loops [13], [14], [15], [16], [17]. Monoclonal antibody trastuzumab is known to bind to the HER2 extracellular domain IV (HER2 domain IV), effectively inhibiting a HER2-mediated cell signaling process [14], [18]. Here we present computationally-guided design and affinity improvement of a protein binder targeting the trastuzumab epitope on domain IV of HER2 which mainly consists of flexible loops. As a model, a protein scaffold composed of LRR (Leucine-rich repeat) modules was employed. We firstly designed protein binders which appear to recognize the target site on domain IV through computational method based on the predicted complex model structures for HER2 domain IV and a protein scaffold. Top 10 designs were expressed, and a lead with a low micromolar binding affinity was selected based on binding and inhibition assays. Binding affinity of the selected lead was further increased by two-orders of magnitude through mutual feedback between computational and experimental approaches. We demonstrated the utility of our approach by determining the binding interface of the developed protein binder through its crystal structure in complex with the HER2 domain IV. Details are reported herein.
2. Materials and methods
2.1. Synthesis and expression of genes
Computationally designed repebody genes and primers used for phage display library were synthesized from Integrated DNA Technologies (Coralville, IA, USA). Synthesized gene fragments went through cloning process after overnight digestion with restriction enzymes (Nde I, Xho I) at 37 °C and ligation (T4 DNA Ligase, Takara Bio, Shiga, Japan) into pET21 vector (Novagen, Madison, WI, USA) at room temperature for 2 h. Materials for bacterial culture were supplied from Duchefa (Haarlem, The Netherlands). Origami B (DE3) competent cells (Novagen) were used for repebody expression. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from LPS Solution (Seoul, Korea). The Ni-NTA agarose resin for purification of his-tagged proteins was purchased from Qiagen (Germantown, MD, USA). Superdex 75 16/600 and Superdex 200 Increase 10/300 size exclusion chromatography columns were purchased from GE Healthcare (Uppsala, Sweden). All other reagents including buffers and solvents were of analytical grade.
2.2. Phage display selection
A repebody library was constructed by overlap PCR using primers containing NNK codon for variable sites on each module. The resulting library was inserted into pBEL118N phagemid [19] and electroporated to TG1 Electroporation-Competent Cells (Agilent Technologies, Santa Clara, CA, USA). Phages containing the library were rescued using M13KO7 Helper Phage (New England Biolabs, Ipswich, MA, USA). Solution-phase bio-panning was conducted in order to minimize the disruption of a target protein. Briefly, 10 μL of Dynabeads M-280 Streptavidin (Invitrogen, Waltham, MA, USA) was loaded into a sterile 1.5 mL centrifuge tube for immobilization of a target protein, and 40 μL of Dynabeads were added to another tube for a negative selection. After washing the beads with PBS (pH.7.4) twice by brief vortex, biotinylated HER2 ectodomain (4 μg/mL, Sino Biological, Beijing, China) was added to the tube and incubated for 2 h at 4 °C. Both HER2 ectodomain-bound beads and negative selection beads were blocked with PBST (PBS pH 7.4, 0.05% Tween 20) containing 2% BSA for 2 h at 4 °C. Phages were prepared in PBST containing in 1% BSA at a final phage concentration of 1.0 × 1012 cfu/mL and mixed with negative selection beads for 1 h. Purified Rb-H0 or Rb-H1 protein was also added to the phage solution in 1 µM concentration for competition. Phage solution was separated from the beads using a magnet and added to HER2 ectodomain-bound beads. After 2 h incubation at room temperature, beads were isolated by using magnetic bar and incubated for 1 min with PBST. This process was repeated five times and finally washed with PBS. To disrupt the binding between a target protein and phage-displayed repebody, 0.2 M glycine-HCl solution (pH 2.2) was added. Beads were isolated by a magnetic bar, and 1 M Tris solution (pH 9.0) was added to the supernatant for neutralization before mixing with TG-1 cells. Phage-infected TG-1 cells were grown in 2xYT agar plate supplemented with 100 μg/mL ampicillin and 1% glucose for overnight at 30 °C. On the following day, 2xYT media were added into the plates to gather the cells and used for next round of selection process. Total five rounds of selection process were conducted for enrichment of positive clones. After the 5th round, cells were diluted at appropriated ratio with PBS (pH 7.4) before plated. On the following day, a 96 deep-well plate (Axygen Scientific, Corning, NY, USA) was seeded with colonies and the resulting phages were acquired for phage ELISA as described in our previous work [20]. Phages were detected by HRP-conjugated anti-M13 antibody (GE Healthcare). For signal generation, 3,3′ 5,5′-tetramethylbenzidine (TMB) (Sigma Aldrich, St. Louis, MO, USA) was used, and the reaction was stopped by addition of 1 N H2SO4. Absorbance at 450 nm was measured with Infinite M200 microplate reader (Tecan, Crailsheim, Germany).
2.3. Computational design and affinity maturation of a site-specific repebody
Initial binding orientations were generated using the ClusPro webserver with the antibody mode [21]. Wild-type repebody (PDB code: 3RFS) was used as a receptor, and the crystal structure of HER2 (1N8Z) was employed as a ligand. The amino acid residues at the convex region were assigned to be repulsive. The residues of the trastuzumab epitope on HER2 domain IV were defined using PyMol from the X-ray crystal structure of trastuzumab in complex with HER2 ectodomain (1N8Z). Any residues on HER2 ectodomain within 5 Å from trastuzumab were defined as epitopes, and attraction was imposed. Rosetta 3.6 (2016.34) with talaris2014 was employed to redesign the binding sites of a repebody toward HER2 domain IV for each binding model [22], [23]. All amino acid types except for cysteine and proline were allowed at each position. In our previous work on the prediction of repebody binding mode [24], the TINKER energy minimization [25] can be used for binding affinity estimation. The AMBER99sb with the GB/SA implicit solvent model was used to minimize the model [26], [27]. For computer-guided affinity maturation, the residues were selected to increase the interaction with HER2 domain IV based on predicted binding modes. Selected residues were randomized to generate a library for phage display, and a clone with highest binding affinity was selected using phage ELISA. The same procedure was repeated to further increase the binding affinity of a selected repebody.
2.4. Enzyme-linked immune-sorbent assay (ELISA)
Binding property of designed and selected repebodies were analyzed by direct ELISA. Briefly, a 96-well Maxibinding plate was coated with extracellular domain of HER2 (10 µg/mL) at 4 °C overnight. PBST containing 1% BSA was used for blocking and dilution of repebodies and antibodies. PBST was used as washing buffer throughout the process. The repebody was detected by using HRP-conjugated anti-c-Myc antibody (1:500 dilution, Santa Cruz Biotechnology, Dallas, TX, USA) or biotinylated anti-repebody antibody (1 μg/mL, AbClon, Seoul, Korea) and HRP-conjugated streptavidin (1:1000 dilution, BioLegend, San Diego, CA, USA). For trastuzumab (Herceptin), HRP-conjugated anti-human Fc antibody (1:10000 dilution, Sigma Aldrich) was used. TMB solution was used for a signal generation and the reaction was stopped using 1 N H2SO4. The signals were measured at 450 nm by microplate reader, and absorbance from maximum concentration was converted to 100% for comparison. For binding specificity test against ErbB family proteins, EGFR, HER3 and HER4 proteins were used (10 µg/mL, Sino Biological).
2.5. Surface plasmon resonance (SPR)
Binding affinity of a repebody was determined through surface plasmon resonance (Biacore T200, GE Healthcare). Briefly, 250 μg/mL of NeutrAvidin Protein (Thermo Scientific, Waltham, USA) was first coated on the surface of CM5 chip (GE Healthcare) in 10 mM sodium acetate buffer (pH 4.5). After immobilization, 20 μg/mL of biotinylated HER2 ectodomain was injected into the chip. Sensorgrams were obtained by flowing a serially diluted repebody into the chip. Kinetic constants were determined by the 1:1 Langmuir binding model using Biacore T200 software (GE Healthcare).
2.6. Size exclusion chromatography for the complex formation
20 μg of HER2 ectodomain from Abcam (Cambridge, UK) was mixed with 5-fold excess amount of a repebody and incubated at 4 °C overnight. The mixtures were injected into Superdex 200 increase 10/300 column for analysis. The peak fractions were analyzed by SDS-PAGE.
2.7. Expression and purification of HER2 domain IV
HER2 domain IV which corresponds to residues from 531 to 626 of HER2 was expressed using insect cells. HER2 gene was subcloned into a baculovirus expression vector by adding Mellitin signal peptide sequences and nona-histidine tag to the N-terminal of HER2 domain IV and TEV cleavage site and maltose-binding protein (MBP) tag to the C-terminal of HER2 domain IV. The expression of HER2 domain IV in insect cells was carried out using a Bac-to-Bac® Baculovirus Expression System (Invitrogen). The resulting construct was expressed in Spodoptera frugiperda (Sf9) insect cells in a secreted form through a culture at 27 °C for 3 days. The media containing secreted HER2 domain IV were collected through centrifugation to remove Sf9 cells and adjusted to a pH of 7.5 for filtration before purification. Next, HER2 domain IV was purified using a HisTrap excel column (GE Healthcare). The filtrated media were loaded into the HisTrap excel column, followed by washing with 20 mM Tris-HCl (pH 7.5), 100 mM NaCl and 20 mM Imidazole. Bound HER2 domain IV was eluted with an elution buffer (20 mM Tris-HCl, pH 7.5) containing 100 mM NaCl and 250 mM Imidazole. Thereafter, the TEV recognition site was cleaved using TEV protease. After desalting to 20 mM Tris-HCl (pH 7.5) containing 50 mM NaCl, HER2 domain IV was loaded into an anion-exchange chromatography column (HiTrap-Q, GE healthcare), and HER2 domain IV was collected from flow through.
2.8. Crystallization, data collection, and structure determination of Rb-H2 in complex with HER2 domain IV
Rb-H2 and HER2 domain IV were mixed at a 1:1.5 M ratio and incubated for 1 h at 4℃. The mixture was applied to the size-exclusion column (HiLoad 16/600 Superdex 200 pg, GE Healthcare). The complex protein between Rb-H2 and HER2 domain IV was concentrated at up to 10 mg/ml and used for crystallization. Initial crystallization screening was conducted by using Mosquito robot (TTP Labtech, Melbourn, UK), and single, appropriate size of crystals appeared at 0.1 M Sodium Citrate: Citric Acid (pH 5.5) and 20% PEG 3000. The complex crystals were quickly soaked into a crystal buffer containing 20% ethylene glycol to protect the crystals from the low temperature of the liquid nitrogen. X-ray diffraction of the complex crystal was then conducted to collect diffraction images using a BL-1A micro-beam line at the Photon Factory (Japan). An integration of the images was conducted using the XDSGUI, and a scaling of the mtz file was also performed using the CCP4 program [28]. The complex crystal belongs to the space group P212121 with a = 44.66 Å, b = 80.07 Å, and c = 108.41 Å in a cell unit. The initial phase was obtained through a molecular replacement (MR) by Molrep using HER2 (PDB ID: 1N8Z) and repebody (PDB ID: 5B4P) as the initial searching model [29]. Model building was conducted using the Coot program, and refinement was carried out using Refmac5 [30], [31]. The three-dimensional model of the complex was visualized using PyMOL.
2.9. Cell culture
Sk-Br3, Sk-Ov3, MCF-7, MDA-MB-468 (ATCC, Manassas, VA, USA) cell lines were cultured in RPMI 1640 media supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin (Capricorn Scientific, Ebsdorfergrund, Germany) at 37 °C incubator with 5% CO2.
2.10. Immunofluorescence labeling and confocal microscopy
NHS-Fluorescein (Thermo Scientific) was prepared in DMSO (Sigma Aldrich) at a concentration of 10 mg/mL and mixed with a repebody dissolved in PBS (pH 7.4) at a dye-to-protein ratio of 10 with a final concentration of a repebody aimed to 2 mg/mL. The mixture of protein and dye was incubated at 4 °C overnight. Excess dye was removed using 0.22 μm centrifugal filter at 13000 rpm for 10 min and subjected to PD-10 desalting column (GE Healthcare). The concentration of a FITC-labeled repebody was measured by NanoDrop 2000c (Thermo Scientific). For confocal microscopy, cells were detached using non-enzymatic cell dissociation solution (Sigma Aldrich) when they reached 80% confluence, and seeded into a 8-well slide glass (SPL Life Sciences) at 3.5x103 cells/well. After 48 h of incubation, cells were gently washed with DPBS (Welgene, Seoul, Korea) for 3 times and treated with a FITC-labeled repebody at 4 °C to prevent endocytosis for 2 h. Following the removal of proteins, cells were gently washed again with DPBS for 3 times and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. After washing with DPBS for three times, cells were stained with DAPI. Cell images were obtained using LSM 780 Confocal Microscopy (Carl Zeiss, Oberkochen, Germany).
2.11. In vitro cytotoxicity
Cells were detached using trypsin-EDTA (Gibco, Waltham, MA, USA) when they reached 80% confluence and seeded into a 96-well plate (SPL Life Sciences) at 1x104 cells/well. After 24 h of incubation, cells were treated with serially diluted (10-fold) repebody or trastuzumab in RPMI 1640 serum free media and further incubated for 72 h at 37 °C and 5% CO2 chamber. Cell cytotoxicity was measured by Cell Counting Kit-8 (Dojindo Molecular Technologies, Kumamoto, Japan). Signals were detected at 450 nm using Infinite M200 microplate reader. Absorbance from cells treated with only media was converted to 100% for comparison.
3. Results
3.1. Computationally-guided design of protein binders targeting a specific site on HER2 domain IV
As proof-of-concept, we aimed to develop a protein binder which targets the trastuzumab epitope on HER2 domain IV and consequently inhibits a HER2-mediated cell signaling. HER2 has no receptor ligands, and triggers cell signaling through homo- or heterodimerization with ErbB protein family [32]. Interestingly, domain IV of all epidermal growth factor receptors is known to consist of structurally flexible loops [16], [18], which may hinder computational design of a protein binder targeting such site. Monoclonal antibody trastuzumab was revealed to bind to the domain IV of HER2, and effectively inhibit a related cell-signaling process [14], [18]. Thus, while extremely challenging, potential therapeutic protein inhibitors should bind to a designated site of HER2 to have expected outcomes, when considering the molecular mechanism of the cell signaling. As a model, a protein scaffold composed of LRR (Leucine-rich repeat) modules, termed ‘Repebody’, was employed. The repebody scaffold showed desirable biochemical and physical properties such as high stability, easy module-based engineering, high bacterial expression, and high tissue penetration [19]. A number of target-specific repebodies have been developed through phage display selection [33], [34], [35]. In particular, the target-binding region of the scaffold is composed of β-strands, exhibiting a rigid-body structure [19].
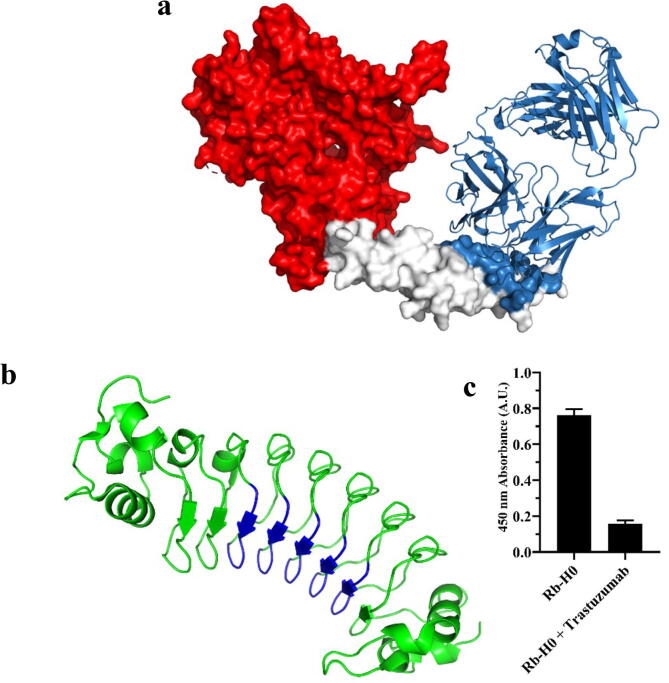
To successfully design a protein binder recognizing a target site through a computational method, the shape complementarity between a protein binder and a target site should be taken into consideration. As the shape complementary of a repebody may not exactly fit the entire trastuzumab epitope on HER2 domain IV, we estimated the chance of the overlap with the trastuzumab epitope by a repebody. Assuming that a library approach generates variants that can bind to any sites of HER2 domain IV, we generated 100,000 random docking models by assigning attraction on two nearby LRR modules, LRRV3 and LRRV4, of the repebody (Fig. 2a). For this, the Rosetta docking protocol was employed [36]. The simulation results show that 100% overlap with the trastuzumab epitope using a repebody may not be possible (Supplementary Fig. S1). In fact, nearly a half of the repebody models are not in contact with the epitope at all. Considering the shape complementarity and steric clashes against trastuzumab for inhibiting the cell signaling, an effective repebody should contain at least > 20% of the trastuzumab epitope. We thus first generated complex model structures using ClusPro [37] with the antibody mode [21] (while not particularly considering the trastuzumab epitope at this stage). Wild-type repebody (PDB ID: 3RFS) was docked onto the target site on domain IV of HER2 (PDB ID: 1N8Z) with repulsion constraints at HER2 domain I-III and the convex region of the repebody (Fig. 1a, b). Total 30 docking models were generated for the target site on the HER2 domain IV. Each LRR module has four variable sites, and 20 variable sites in total on five modules of wild-type repebody were subjected to redesign based on the docking models using RosettaScript protocol [22]. Only domain IV was considered in the Rosetta redesign process, and one thousand designs were generated for each docking model. Both proline and cysteine were excluded in the design process. Among the 30,000 designs, top 10 clones with the lowest energy values and those which appeared to share > 20% of the trastuzumab epitope were selected. It should be noted that one of the initial docking models that share the epitope is very similar to the crystal structure of Rb-H2 in complex with the domain IV (Supplementary Fig. S2). However, this docking model was different from the one chosen from the experimental result, which is actually used in the further affinity maturation process since the purpose of our study was to design a protein binder that targets a specific epitope, not a binder with certain binding mode. The selected clones were expressed in E. coli and subjected to purification, followed by binding assays using ELISA against the HER2 ectodomain (Supplementary Fig. S3). We finally selected Rb-H0 as a lead showing the highest binding signal to HER2 ectodomain. There was a 5-fold signal decrease in the presence of trastuzumab (Fig. 1c), which indicates that Rb-H0 shares the trastuzumab epitope. To estimate the binding affinity of Rb-H0, we analyzed the binding profile against HER2 using direct ELISA with the increasing concentration of Rb-H0. As a result, the binding affinity of Rb-H0 was estimated to be 4 μM (Fig. 2b, Supplementary Table S1).
Fig. 2.
Computationally-guided affinity improvement of Rb-H0 and biophysical properties of affinity-maturated Rb-H2. a, Modules used in the first and second round of affinity maturation of Rb-H0 are shown in representative structure of a repebody scaffold. Annotation of the modules is indicated, and each module is numbered from N-terminus to C-terminus. Seven residues in modules LRRV3 and LRRV4 were used for first-round affinity maturation. Additional seven residues on modules LRR1, LRRV1, and LRRV2 were optimized for second round affinity maturation. b, Binding profiles of Rb-H0 and affinity-maturated Rb-H1 and Rb-H2 by ELISA. Error bars represent average ± standard deviation (n = 3). c, Determination of binding affinity of Rb-H2 for HER2 ectodomain through surface plasmon resonance (SPR).
Fig. 1.
Selection of Rb-H0 binding to HER2 domain IV from the computationally designed protein binders. a, Initial docking models were generated on HER2 domain IV with assigned repulsion on domain I-III (red). Trastuzumab and its epitope are colored in skyblue. b, Non-concave region of a repebody model (PDB ID: 3RFS chain A) was masked (blue). c, Selection of Rb-H0 from the computationally designed candidates. Among top 10 designs showing the lowest energy levels, Rb-H0 exhibiting the highest signal and significant decrease in the signal in the presence of trastuzumab was selected as the initial binder. Error bars represent average ± standard deviation (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Affinity improvement by an integrated computational and experimental approach
Since Rb-H0 has a low binding affinity for HER2 ectodomain, we intended to increase its binding affinity. Based on the docking models of wild-type repebody against HER2 domain IV, we reasoned that the residues at modules LRRV3 and LRRV4 on Rb-H0 would have the highest proximity for the targeted site on HER2 domain IV. Seven residues (Ile114, Asp116, Ser118, Asn119, Ile138, Asp140, Ser142) were selected and randomized for a library construction followed by phage display selection (Fig. 2a). A clone with the highest binding signal, designated as Rb-H1, was shown to have a significantly increased binding affinity compared with Rb-H0 (Fig. 2b, Supplementary Table S1). For the second round of affinity maturation, we predicted the binding mode of Rb-H1 to the HER2 domain IV using the computational method as described elsewhere [24]. It should be noted that the computational binding mode prediction requires not only computational energy scores but also solid experimental validation in advance, such as paratope information and clues on epitopes by mutagenesis. Thus, binding mode prediction for Rb-H0 was not carried out since it lacks in such information. In the docking process, ClusPro with the antibody mode was employed for protein docking [21], [37]. Rb-H1 structure was modeled based on wild-type repebody structure, and repulsion was assigned on the convex residues. Attraction was imposed on the library sites for affinity improvement. As it was known from the competitive binding assay that Rb-H1 might share the epitope with trastuzumab, any docking models that were not in contact with the trastuzumab epitope were eliminated. Total 17 docking models were finally selected, followed by energy-minimization. The docking model with the lowest energy was predicted to be the binding mode of Rb-H1 [24] (Supplementary Fig. S4). Based on the predicted binding mode, another seven residues (Gln46, Ile48, Asn50, Asn51, Tyr68, Ala70, Val90) on the three modules LRR1, LRRV1 and LRRV2, at the N-terminus were chosen and randomized for a library construction and phage display selection (Fig. 2a). As a result, a variant with the highest signal, Rb-H2, was selected, and it was observed to have a marginal increase in binding affinity compared with Rb-H1 (Fig. 2b, Supplementary Table S1). The binding affinity of Rb-H2 was determined to be 54 nM through surface plasmon resonance (SPR) (Fig. 2c) which is a significant increase in the binding affinity of a lead protein binder by two-orders of magnitude.
After the second-round affinity-maturation, however, only a marginal increase in the binding affinity of Rb-H2 was observed despite seven additional mutations. Amino acid sequence of Rb-H0, Rb-H1, and Rb-H2 are shown in Supplementary Table S2. Given that information, we assumed that only certain mutations would contribute to the increase in the binding affinity of Rb-H1. We analyzed the binding energy for each single mutant of Rb-H2 based on the model complex for Rb-H1 (Supplementary Table S3). The energy calculation results indicate that two single mutations (V90T and N51H) make significant contributions to the increase in the binding affinity. By taking into account the calculation results, we remodeled the binding mode of Rb-H2 again by assigning attractions at the two predicted positions (V90T and N51H). After energy minimization, the final model was shown to be well coincident with the X-ray crystal structure (I-RMSD: 1.701 Å, fnat: 0.508, Supplementary Fig. S4).
To further get insight into the binding site and affinity of the variants, competitive ELISA was carried out for Rb-H0, Rb-H1, and Rb-H2 in the presence of trastuzumab (Fig. 3a). The signals of the variants were shown to decrease in the presence of trastuzumab, indicating that they share the trastuzumab epitope as intended and predicted. In the case of Rb-H2, the signal decrease was much smaller compared to Rb-H0, supporting a significant increase in binding affinity, considering the binding affinity of trastuzumab for HER2 domain IV (5 nM [38]). We tested the specificity of Rb-H2 against ErbB family proteins with high structural similarity to HER2, including EGFR, HER2, HER3, and HER4. As a result, Rb-H2 showed the highest binding specificity for HER2 ectodomain (Fig. 3b). Finally, we checked whether Rb-H2 forms a complex with HER2 ectodomain using size exclusion chromatography (Fig. 3c). Rb-H2 in complex with HER2 ectodomain was eluted as a single peak at a designated position, confirming the complex formation between Rb-H2 and HER2 ectodomain. In addition, we double confirmed the complex formation between Rb-H2 and HER2 domain IV in gel filtration chromatography (Data not shown). The results support that Rb-H2 indeed shares a binding site on HER2 domain IV with trastuzumab as intended.
Fig. 3.
Characteristics of affinity-maturated Rb-H2. a, Competitive inhibition assays of Rb-H0, Rb-H1 and Rb-H2 in the presence of trastuzumab (1 mg/mL). All the binders showed decreased signals in the presence of trastuzumab. Error bars represent average ± standard deviation (n = 3). b, Specificity of Rb-H2 against ErbB family proteins. Rb-H2 was able to distinguish HER2 among the ErbB family proteins. Error bars represent average ± standard deviation (n = 3). c, Complex formation between Rb-H2 and HER2 ectodomain. Two proteins were mixed and eluted through size exclusion chromatography. Two proteins were eluted together in the first fraction as shown in SDS-PAGE (inset).
3.3. X-ray crystal structure of Rb-H2 in complex with HER2 domain IV
To confirm the binding site of Rb-H2, we determined the X-ray crystal structure of Rb-H2 in complex with HER2 domain IV at 2.03 Å resolution (Fig. 4a, c). The crystallographic and refinement statistics are shown in Supplementary Table S4. Rb-H2 is shown to bind to the targeted site of HER2 domain IV containing the trastuzumab epitope. The interface area between Rb-H2 and HER2 domain IV was estimated to be 2,070 Å2, whereas the interface area of trastuzumab is about 1,958 Å2. The binding site of Rb-H2 overlaps with that of the trastuzumab, covering approximately one-fourth of the trastuzumab epitope, which resulted in the binding competition against trastuzumab. Structural analysis revealed that HER2 domain IV interacts primarily with the concave side of Rb-H2 through hydrophobic interactions, hydrogen bonds and salt-bridge. Thr137 of Rb-H2 forms hydrogen bond with Gly572 of HER2 domain IV, and Trp116 and Arg140 of Rb-H2 have hydrophobic interactions with Val546, Leu547 and Val574 of HER2 domain IV (Fig. 4e). In addition, Arg240 of Rb-H2 forms a salt bridge with Asp582 of HER2 domain IV, and four residues (Lys185, Tyr210, Arg240 and Gly244) of Rb-H2 have hydrogen bonds with four residues (Val585, Cys584, Asp582 and Gln583) of HER2 domain IV, respectively (Fig. 4f). Specifically, three residues (Ser48, His51 and Tyr70) of Rb-H2 form hydrogen bonds with three residues (Glu543, Gln533, and Gln548) of HER2 domain IV (Fig. 4d). Val162, Tyr210 and Trp212 of Rb-H2 are shown to have hydrophobic interactions with Ser573, Phe577 of HER2 domain IV (Fig. 4e). It is interesting to note that some amino acid residues of Rb-H2 mentioned above (Ser48, His51, Tyr70, Trp116 and Arg140) are changed from those of Rb-H0. Overall, our structural analysis supports the significantly improved binding affinity of Rb-H2 for HER2 domain IV by two-orders of magnitude. Furthermore, based on the structure of HER2 domain IV, the binding region of Rb-H2 overlaps with the epitope of trastuzumab as shown in Fig. 4a and b. The binding surface areas of HER2 domain IV/Rb-H2, HER2 domain IV/ trastuzumab and the overlapped area are as follows: 2,070 Å2, 1,958 Å2 and 481 Å2 (23% of the trastuzumab epitope), respectively. The overlapped residues between HER2 domain IV/Rb-H2 and HER2 domain IV/trastuzumab include Phe577, Asp582, Gln583 and Lys591 of HER2 (Fig. 4b) [15]. Based on our initial docking simulation (Supplementary Fig. S1), the probability that a repebody generated from a random library approach shares > 23% of the trastuzumab epitope is 0.3. The results provide distinct insight into the utility and potential of our computational-driven design approach.
Fig. 4.
Crystal structure of Rb-H2 in complex with HER2 domain IV. a, Overall structure of Rb-H2 in complex with HER2 domain IV. The complex structure of trastuzumab in complex with HER2 domain IV is also shown. HER2 domain IV is presented in surface model, and Rb-H2 and trastuzumab are in cartoon model. Two structures are superimposed based on HER2 Domain IV. (HER2 domain IV: bluewhite, Rb-H2: warmpink, trastuzumab: skyblue, overlapped: dirtyviolet). b, Binding regions on HER2 domain IV of Rb-H2 and trastuzumab. The overlapped residues are marked. c, Overall structure of Rb-H2 in complex with HER2 domain IV. Rb-H2 and HER2 domain IV are presented in cartoon model. Three interaction regions are represented in detail in d-f. d, Hydrogen bonds are shown in dashed lines in stick model. e, Hydrogen bonds are shown in dashed lines in stick model and hydrophobic interaction residues are presented in stick model. f, Hydrogen bonds and salt-bridge are shown in dashed lines in stick model.
3.4. In vitro binding and cytotoxicity of Rb-H2
We examined the binding of the developed Rb-H2 to HER2 on the cell surface. For this, cancer cell lines expressing different levels of HER2 were tested, including Sk-Br3 (high expression), Sk-Ov3 (moderate), and MCF-7 (low). We labeled Rb-H2 with fluorescein isothiocyanate (FITC) and treated it with the cells followed by imaging using confocal microscope. As shown in Fig. 5a, the strong fluorescence intensity was observed on the peripheral region of Sk-Br3 cells, whereas MCF-7 cells exhibited the lowest fluorescent intensity. Based on the result, it is evident that Rb-H2 binds to HER2 ectodomain on the cell surface. No fluorescence was detected when an off-target repebody (human serum albumin specific repebody [20]) labeled with fluorescein was treated with each cell line, supporting the specific binding of Rb-H2 to HER2 ectodomain on the cell surface.
Fig. 5.
Binding of Rb-H2 to HER2-expressing cells and its cytotoxicity in vitro. a, Confocal microscopic images of HER2-expressing cancer cell lines after treatment with fluorescein-labeled Rb-H2. Cells were treated with 1 μM of labeled Rb-H2 for 3 h at 37 °C and imaged by confocal microscopy. Sk-Br3 (high level of HER2 expression, top), Sk-Ov3 (moderate HER2 level, middle), and MCF (low HER2 expression, down) cells were used. b, In vitro cytotoxicity of Rb-H2. Cells expressing HER2 were treated with Rb-H2 or trastuzumab at different concentrations for 72 h, and cell viability was determined by CCK-8 assay. Error bars represent average ± standard deviation (n = 3).
Since Rb-H2 was developed by targeting the trastuzumab epitope on HER2 domain IV, it is expected to inhibit the HER2-mediated cell signaling as trastuzumab does. We tested the cytotoxicity of Rb-H2 for various cancer cell lines (Fig. 5b). In the case of Sk-Br3, cell viability gradually decreased with the increasing concentration of Rb-H2 and reached 70% at the concentration of 1 μM. Similar cytotoxicity was observed for Sk-Ov3 and MCF-7, even though their HER2 expression levels were lower than Sk-Br3. Rb-H2 was shown to exhibit a similar cytotoxic pattern to trastuzumab for the tested cell lines, implying that it inhibits the cell signaling process in a similar way to trastuzumab because it shares the binding site with trastuzumab. Trastuzumab is clinically used for treatment of breast cancer and showed a saturation pattern even at high concentration. It is evident that Rb-H2 binds to the targeted site of HER2 domain IV and consequently inhibits the cell signaling pathway as trastuzumab by blocking dimerization, suppressing the cell proliferation.
4. Discussion
We demonstrated computationally-guided design and affinity improvement of a protein binder recognizing a specific site on domain IV of HER2. Rational design of a protein binder with a desired epitope and binding affinity has been a long-standing goal in protein engineering field. Our strategy involves the computational design of protein binders which appeared to recognize a target site, followed by selection of potentially effective binders through experimental binding assays. As proof-of-concept, we aimed to design a protein binder which target the trastuzumab epitope on HER2 domain IV. The domain IV has very little content of secondary structures, which is supposed to be “non-ideal” features to be targeted by computational design approach. It has been shown that high flexibility of the domain makes it even harder to computationally design a protein binder recognizing such domain [12]. Nonetheless, our approach enabled a successful design of a protein binder recognizing a target site on domain IV of HER2 as intended.
Epitope and binding affinity of a protein binder is crucial for its therapeutic efficacy. Development of a protein binder with a desirable epitope and binding affinity has mostly relied on experimental approaches comprising repeated rounds of a library construction and screening, but they are labor-intensive and difficult to identify the binding epitope during experiments. Recently, computational methods have shown notable successes in the design of proteins with desired functions due to many advances in the computing power and algorithms. However, purely computational design of a protein binder targeting a specific site still remains a challenge mainly because the current computational energy scoring is not accurate enough to precisely predict the binding free energy landscapes and may not be generally applicable. Furthermore, if a target protein is composed of multi-domains like extracellular receptors, computational design of such protein binders becomes extremely difficult. Our computationally-guided approach effectively generated the protein binder candidates for each docking model between a protein scaffold and HER2 domain IV by taking into consideration of shape complementarity. The docking models showed that a perfect overlap with the trastuzumab epitope using a repebody would be impossible as expected. Considering the shape complementarity and steric clashes against trastuzumab for inhibiting the cell signaling, we reasoned that an effective design should share at least > 20% of the trastuzumab epitope. With this criterion, total 30 docking models were generated for the target site on the HER2 domain IV, and 20 variable sites of wild-type repebody were computationally redesigned for each model using RosettaScript protocol. Our approach eventually enabled a lead with a low micromolar affinity among the 30,000 designs. Based on the results, it is likely that shape complementarity is critical to the design of protein binder recognizing a target site. In addition, our combined approach was shown to be effective for significantly enhancing the affinity of an initial binder, proving its utility for affinity improvement.
The X-ray crystal structure of Rb-H2 in complex with HER2 domain IV validated the utility of our approach by confirming that Rb-H2 indeed binds to the target site on HER2 domain IV as intended, showing the overlap (approximately one fourth) with the trastuzumab epitope. It is interesting to note that the computationally predicted binding orientation of Rb-H2 against HER2 domain IV was well coincident with the X-ray crystal structure, which supports the utility of the computational method to model the binding mode [24]. Binding of Rb-H2 to the HER2-expressing cells and its in vitro cytotoxicity also supported the potential of our approach.
Taken together, the present strategy can be widely applied to the development of a protein binder with a desired epitope and binding affinity for a target protein as an alternative to conventional experimental methods.
CRediT authorship contribution statement
Tae Yoon Kim: Conceptualization, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Jeong Seok Cha: Validation, Formal analysis, Investigation, Writing - original draft. Hoyoung Kim: Formal analysis, Investigation. Yoonjoo Choi: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Funding acquisition. Hyun-Soo Cho: Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition. Hak-Sung Kim: Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition. : .
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program (NRF-2017M3A9F5031419 to H.-S.K., NRF-2017M3A9F6029755, NRF-2019M3E5D6063903 to H.-S.C), Global Research Laboratory (NRF-2015K1A1A2033346 to H.-S.K.), Mid-Career Researcher Program (NRF-2017R1A2A1A05001091 to H.-S.K.), NRF-2018R1A5A2024181 to Y.j.C., Science Research Center (NRF-2016R1A5A1010764 to H.-S.C) of the National Research Foundation (NRF) funded by the Ministry of Science and ICT of Korea. We thank the staff scientists for assistance at the beamline 1A and 17A of the Photon Factory and the beamline 11C of Pohang Light Source.
Data availability
Protein structure information are deposited in Protein Data Bank (Accession code: 6LBX).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.02.013.
Contributor Information
Yoonjoo Choi, Email: kalicuta@gmail.com.
Hyun-Soo Cho, Email: hscho8@yonsei.ac.kr.
Hak-Sung Kim, Email: hskim76@kaist.ac.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ledford H. Monoclonal antibodies come of age. Nature. 2008;455:437. doi: 10.1038/455437a. [DOI] [PubMed] [Google Scholar]
- 2.Chames P., Van Regenmortel M., Weiss E., Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerner R.A. Manufacturing immunity to disease in a test tube: the magic bullet realized. Angew Chem Int Ed Engl. 2006;45(48):8106–8125. doi: 10.1002/anie.200603381. [DOI] [PubMed] [Google Scholar]
- 4.Dunn IS. Searching for molecular solutions: empirical discovery and its future. Hoboken, N.J.: John Wiley & Sons; 2010.
- 5.Silva D.A., Yu S., Ulge U.Y., Spangler J.B., Jude K.M., Labao-Almeida C. De novo design of potent and selective mimics of IL-2 and IL-15. Nature. 2019;565:186–191. doi: 10.1038/s41586-018-0830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevalier A., Silva D.A., Rocklin G.J., Hicks D.R., Vergara R., Murapa P. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550:74–79. doi: 10.1038/nature23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramisch S., Weininger U., Martinsson J., Akke M., Andre I. Computational design of a leucine-rich repeat protein with a predefined geometry. Proc Natl Acad Sci U S A. 2014;111:17875–17880. doi: 10.1073/pnas.1413638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tinberg C.E., Khare S.D., Dou J., Doyle L., Nelson J.W., Schena A. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleishman S.J., Whitehead T.A., Ekiert D.C., Dreyfus C., Corn J.E., Strauch E.M. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon D.A., Shan L., Du Q., Shirinian L., Rickert K.W., Rosenthal K.L., Korade M., 3rd, van Vlerken-Ysla L.E., Buchanan A., Vaughan T.J., Damschroder M.M., Popovic B. Experimentally guided computational antibody affinity maturation with de novo docking, modelling and rational design. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houk K.N., Liu F. Holy grails for computational organic chemistry and biochemistry. Acc Chem Res. 2017;50:539–543. doi: 10.1021/acs.accounts.6b00532. [DOI] [PubMed] [Google Scholar]
- 12.Whitehead T.A., Baker D., Fleishman S.J. Computational design of novel protein binders and experimental affinity maturation. Methods Enzymol. 2013;523:1–19. doi: 10.1016/B978-0-12-394292-0.00001-1. [DOI] [PubMed] [Google Scholar]
- 13.Menard S., Pupa S.M., Campiglio M., Tagliabue E. Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003;22:6570–6578. doi: 10.1038/sj.onc.1206779. [DOI] [PubMed] [Google Scholar]
- 14.Tebbutt N., Pedersen M.W., Johns T.G. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer. 2013;13(9):663–673. doi: 10.1038/nrc3559. [DOI] [PubMed] [Google Scholar]
- 15.Cho H.S., Mason K., Ramyar K.X., Stanley A.M., Gabelli S.B., Denney D.W., Jr. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 16.Banappagari S., Ronald S., Satyanarayanajois S.D. A conformationally constrained peptidomimetic binds to the extracellular region of HER2 protein. J Biomol Struct Dyn. 2010;28:289–308. doi: 10.1080/07391102.2010.10507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastner J., Loeffler H.H., Roberts S.K., Martin-Fernandez M.L., Winn M.D. Ectodomain orientation, conformational plasticity and oligomerization of ErbB1 receptors investigated by molecular dynamics. J Struct Biol. 2009;167:117–128. doi: 10.1016/j.jsb.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Arkhipov A., Shan Y., Kim E.T., Dror R.O., Shaw D.E. Her2 activation mechanism reflects evolutionary preservation of asymmetric ectodomain dimers in the human EGFR family. Elife. 2013;2 doi: 10.7554/eLife.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S.C., Park K., Han J., Lee J.J., Kim H.J., Hong S. Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering. Proc Natl Acad Sci U S A. 2012;109:3299–3304. doi: 10.1073/pnas.1113193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T.Y., Park J.H., Shim H.E., Choi D.S., Lee D.E., Song J.J. Prolonged half-life of small-sized therapeutic protein using serum albumin-specific protein binder. J Control Release. 2019;315:31–39. doi: 10.1016/j.jconrel.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Brenke R., Hall D.R., Chuang G.Y., Comeau S.R., Bohnuud T., Beglov D. Application of asymmetric statistical potentials to antibody-protein docking. Bioinformatics. 2012;28:2608–2614. doi: 10.1093/bioinformatics/bts493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleishman S.J., Leaver-Fay A., Corn J.E., Strauch E.M., Khare S.D., Koga N. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Meara M.J., Leaver-Fay A., Tyka M.D., Stein A., Houlihan K., DiMaio F. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J Chem Theory Comput. 2015;11:609–622. doi: 10.1021/ct500864r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y., Jeong S., Choi J.M., Ndong C., Griswold K.E., Bailey-Kellogg C., Kim H.S. Computer-guided binding mode identification and affinity improvement of an LRR protein binder without structure determination. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rackers J.A., Wang Z., Lu C., Laury M.L., Lagardère L., Schnieders M.J. Tinker 8: software tools for molecular design. J Chem Theory Comput. 2018;14(10):5273–5289. doi: 10.1021/acs.jctc.8b00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Still W.C., Tempczyk A., Hawley R.C., Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 28.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin A.A., Isupov M.N. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr. 2001;57:1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Baselga J., Swain S.M. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9(7):463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.J., Kim H.J., Yang C.S., Kyeong H.H., Choi J.M., Hwang D.E. A high-affinity protein binder that blocks the IL-6/STAT3 signaling pathway effectively suppresses non-small cell lung cancer. Mol Ther. 2014;22:1254–1265. doi: 10.1038/mt.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.J., Choi H.J., Yun M., Kang Y., Jung J.E., Ryu Y. Enzymatic prenylation and oxime ligation for the synthesis of stable and homogeneous protein-drug conjugates for targeted therapy. Angew Chem Int Ed Engl. 2015;54:12020–12024. doi: 10.1002/anie.201505964. [DOI] [PubMed] [Google Scholar]
- 35.Hwang D.E., Ryou J.H., Oh J.R., Han J.W., Park T.K., Kim H.S. Anti-human VEGF repebody effectively suppresses choroidal neovascularization and vascular leakage. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitzner B.D., Jeliazkov J.R., Lyskov S., Marze N., Kuroda D., Frick R. Modeling and docking of antibody structures with Rosetta. Nat Protoc. 2017;12:401–416. doi: 10.1038/nprot.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herceptin (Trastuzumab) [package insert]. U.S. Food and Drug Administration; 1998.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein structure information are deposited in Protein Data Bank (Accession code: 6LBX).