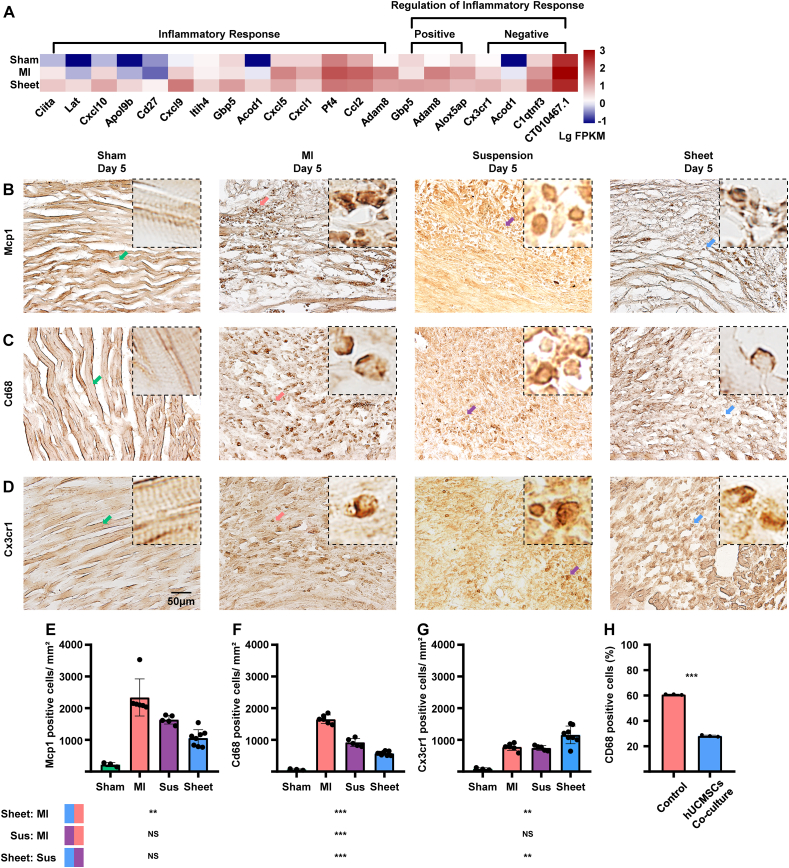
Fig. 3.
Evaluation of inflammatory effects in the ischemic heart injury area of ICR mice. (A)Transcriptome array and GO:BP analysis with inflammatory response-related genes (n = 2, each group). (B) IHC staining of treatment and MI-only groups 5 days post-MI with monocyte marker Mcp1. Scale bar = 50 μm (C)IHC staining of treatment and MI-only groups 5 days post-MI for the M1 macrophage marker Cd68. Scale bar = 50 μm. (D) IHC staining of treatment and MI-only groups 5 days post-MI with non-classical macrophagemarker Cx3cr1. Scale bar = 50 μm. (E–G) Quantitative analysis of Mcp1-, Cd68-and Cx3cr1-positive cells/mm2; values are mean ± SD (control group, n = 3; MI-only group, n = 6; suspension groups, n = 5; cell sheet group,n = 8). NSCell sheet group (blue) < cell suspension group (purple) with Mcp1-positive cells (P > 0.05); Kruskal–Wallis H test with Bonferroni correction (E); ***cell sheet group (blue) < cell suspension group (purple) withCD68-positive cells (P < 0.001); **cell sheet group (blue) < cell suspension group (purple) with Cx3cr1-positive cells (P < 0.01) by one-way ANOVA with Tukey comparison test (F, G), ***hUCMSCs co-cultured group >control group with CD68-positive cells rate (P < 0.001); t-test. GO:BP, Gene Ontology Biological Process; IHC, immunohistochemistry; MI, myocardial infarction; Sus, suspension; Mcp1, monocyte chemoattractant protein 1; SD, standard deviation; ANOVA, analysis of variance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)