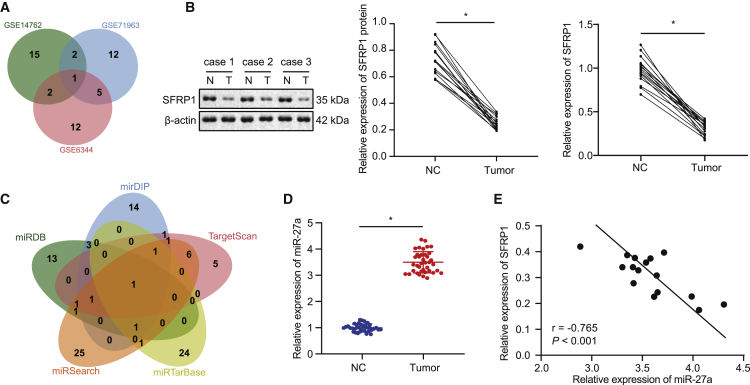
Figure 1.
SFRP1 was poorly expressed and miR-27a was highly expressed in RCCC, showing a negative correlation
(A) Comparison of the top 20 differentially expressed genes in RCCC-related profiles GSE14762, GSE71963, and GSE6344, with only one intersection of the SFRP1 gene. (B) Western blot analysis of SFRP1 protein levels in RCCC tissues and adjacent tissues (left panel, middle panel), and qRT-PCR analysis of SFRP1 mRNA levels normalized to GAPDH (right panel) (N = 16). (C) The jvenn method used to compare miRDB, mirDIP, TargetScan, miRTarBase, and miRSearch databases to predict specific miRNAs targeting SFRP1, where there was only one intersection of miRNA: hsa-miR-27a. (D) qRT-PCR analysis of miR-27a expression normalized to U6 in RCCC tissues and adjacent tissues (N = 38). (E) Pearson’s correlation analysis of miR-27a and SFRP1 mRNA expression in RCCC tissues (N = 16); ∗p < 0.05 versus adjacent tissues. The measurement data are summarized by mean ± SD. Cancer tissues and adjacent non-cancerous tissues were analyzed by paired t test. Pearson coefficient was employed to evaluate the correlation between miR-27a and SFRP1.