Figure 5. The t-SNARE SNAP47 interacts with TRIM67 and localizes to VAMP2-mediated exocytic events.
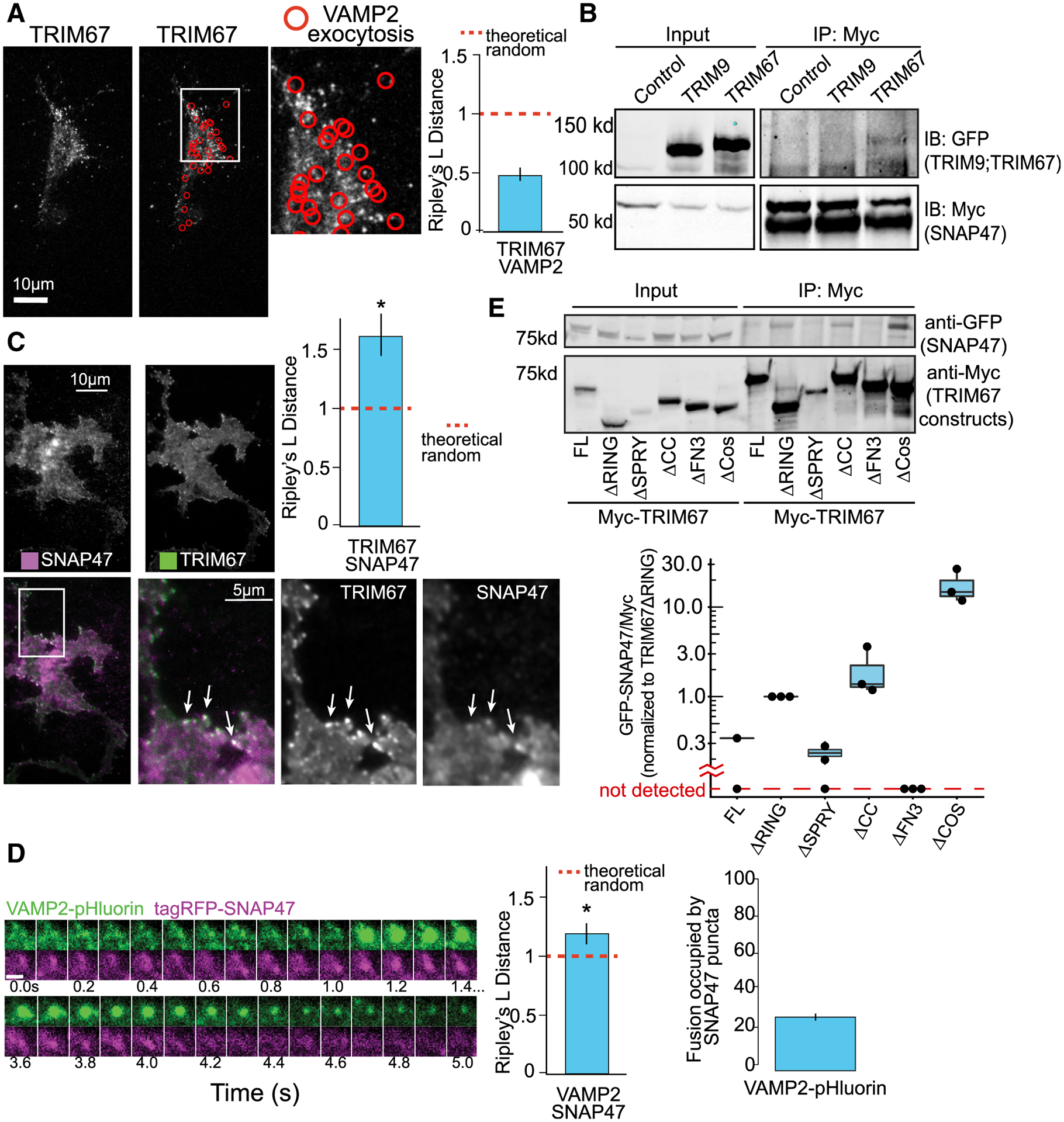
(A) Representative images and Ripley’s L distance +/− standard error colocalization analysis of TRIM67-tagRFP and VAMP2-pHluorin events (red circles) n = 15 cells; n = 4 biological replicates; value lower than theoretical random suggests no colocalization.
(B) Immunoprecipitation (IP:Myc) of Myc-SNAP47 from HEK293 cells expressing Myc-SNAP47 and either GFP-TRIM67 or GFP-TRIM9, blotted for GFP and Myc.
(C) Average of 10 frames of images and colocalization analysis of TagRFP-SNAP47 and TRIM67-GFP in cortical neurons at 2 DIV (n = 14 cells; n = 3 biological replicates; values above theoretical line are colocalized (*). Error bars report standard error.
(D) Example of VAMP2-pHluorin and TagRFP-SNAP47 colocalization during an exocytic event. n = 16 cells; values above theoretical line are colocalized (*). Error bars report standard error. Scale bar, 1 μm.
(E) Structure-function coimmunoprecipitation (coIP) assays. MycTRIM67 construct IP blotted for GFP-SNAP47 and Myc. Graph represents relative GFP-SNAP47/Myc-TRIM67 constructs normalized to Myc-TRIM67ΔRING; blots without a detectable band of GFP-SNAP47 represented below graph break.
