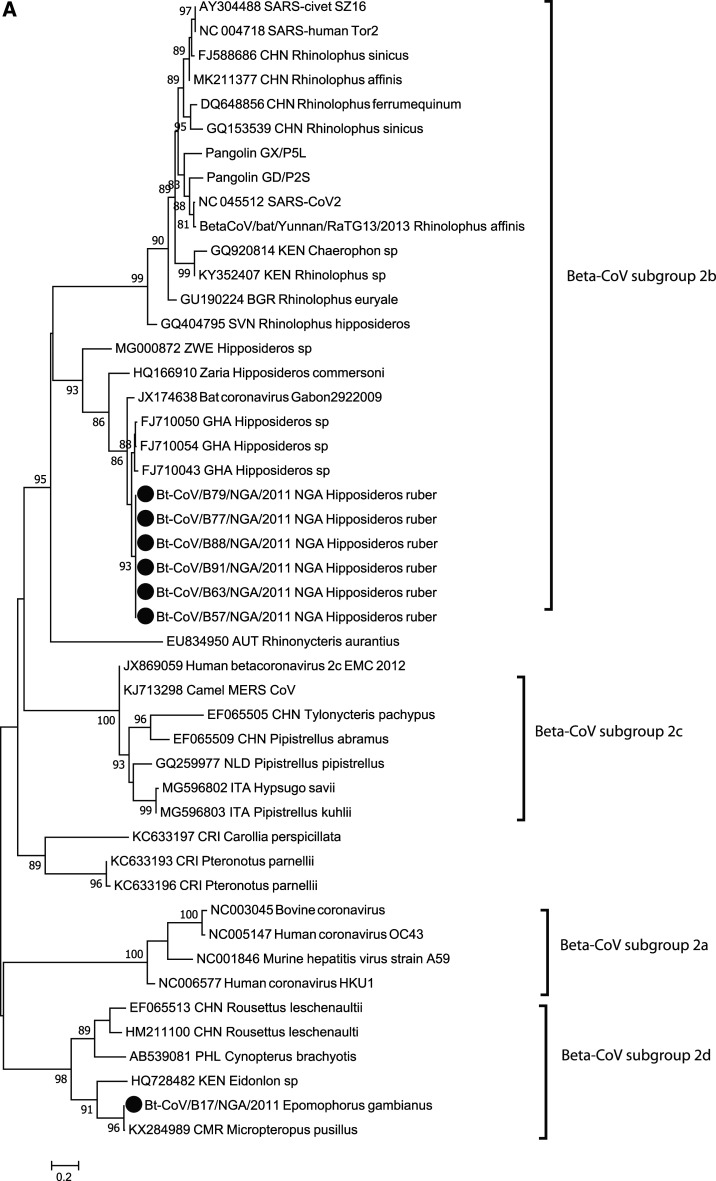
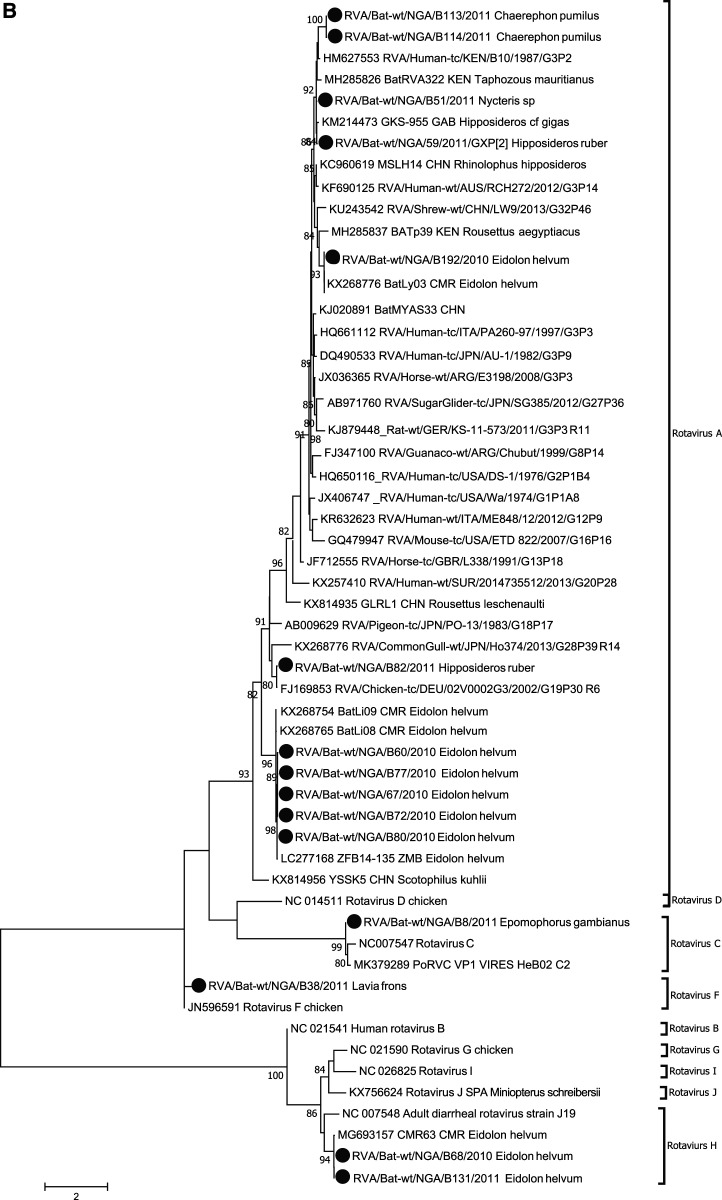
Figure 1.
Phylogenetic analysis of bat betacoronavirus (A) and bat rotavirus (RV) (B) from Nigeria. Phylogenetic trees were constructed using the maximum likelihood (ML) method available PhyML version 3.0 assuming a general time-reversible model with a discrete gamma distributed rate variation among sites (G4) and a subtree prunning and regrafting tree swapping algorithm. The seven Nigeria bat betacoronaviruses and 15 bat RVs are highlighted with solid circles. The betacoronavirus subgroup and RV species information is shown to the right side of the phylogeny. The maximum likelihood bootstrap is indicated next to the nodes. The scale bar indicates the estimated number of nucleotide substitutions per site.