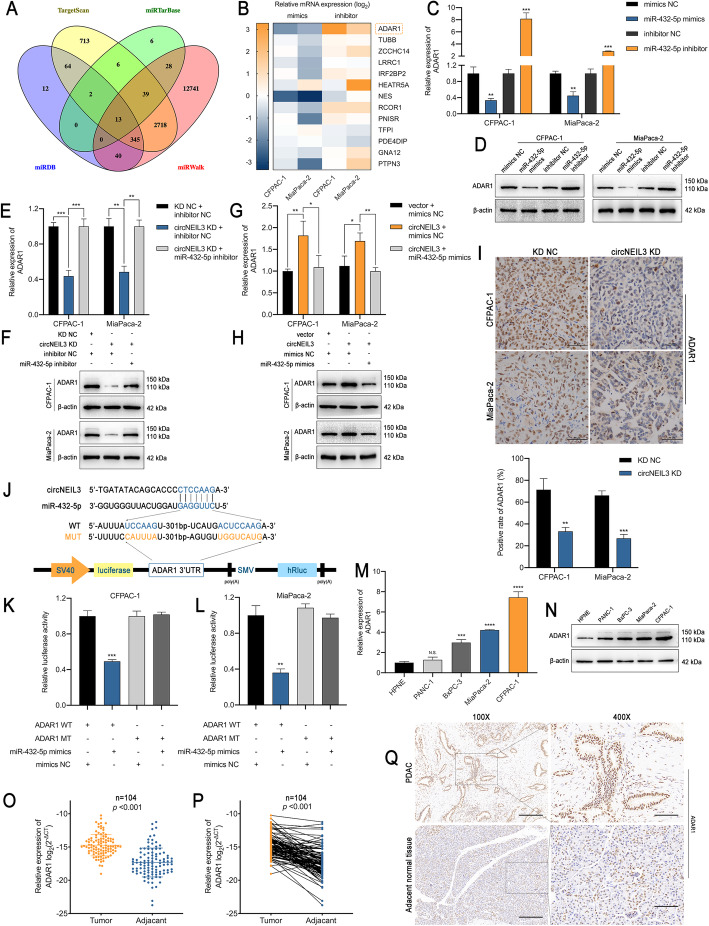
Fig. 6.
ADAR1 is a direct target of miR-432-5p and is indirectly regulated by circNEIL3 a. Venn diagram showing 13 genes that are putative miR-432-5p targets predicted by four algorithms (Targetscan, miRTarBase, miRDB and miRWalk). b. A heat map was used to visualize the expression of the predicted target genes in CFPAC-1 and MiaPaca-2 cells transfected with the miR-432-5p mimics, inhibitor and corresponding NC. c-d. ADAR1 expression in CFPAC-1 and MiaPaca-2 cells was analyzed by RT-qPCR and western blot analyses after transfection with the miR-432-5p mimics or inhibitor. e-h. ADAR1 expression was analyzed by RT-qPCR and western blot analyses. CFPAC-1 and MiaPaca-2 cells were transfected with the miR-432-5p mimic or cotransfected with the indicated circNEIL3 vectors. i. IHC staining of xenograft tumours. The protein levels of ADAR1 were analyzed based on IHC staining. The samples were imaged at 400× magnification. Scale bar = 50 μm. j. Schematic of the ADAR1 3’UTR wild-type (WT) and mutant (MUT) luciferase reporter vectors. k-l. The relative luciferase activities were analyzed in CFPAC-1 and MiaPaca-2 cells cotransfected with the miR-432-5p mimics or mimics NC and the ADAR1 3’UTR wild-type (WT) or mutant (MUT) luciferase reporter vectors. m-n. RT-qPCR and western blot analyses of the relative expression levels of ADAR1 in HPNE, PANC-1, BxPC-3, MiaPaca-2 and CFPAC-1. o-p. RT-qPCR analysis of ADAR1 expression in PDAC tissues (n = 104) paired with adjacent normal tissues (n = 104). q. IHC staining of ADAR1 of PDAC and adjacent normal tissues from patients. The samples were imaged at 100× and 400× magnification. Scale bar = 100 and 25 μm. All data are presented as the means ± SD of three independent experiments. n.s., no significance; *p < 0.05, **p < 0.01, ***p < 0.001