Figure 4.
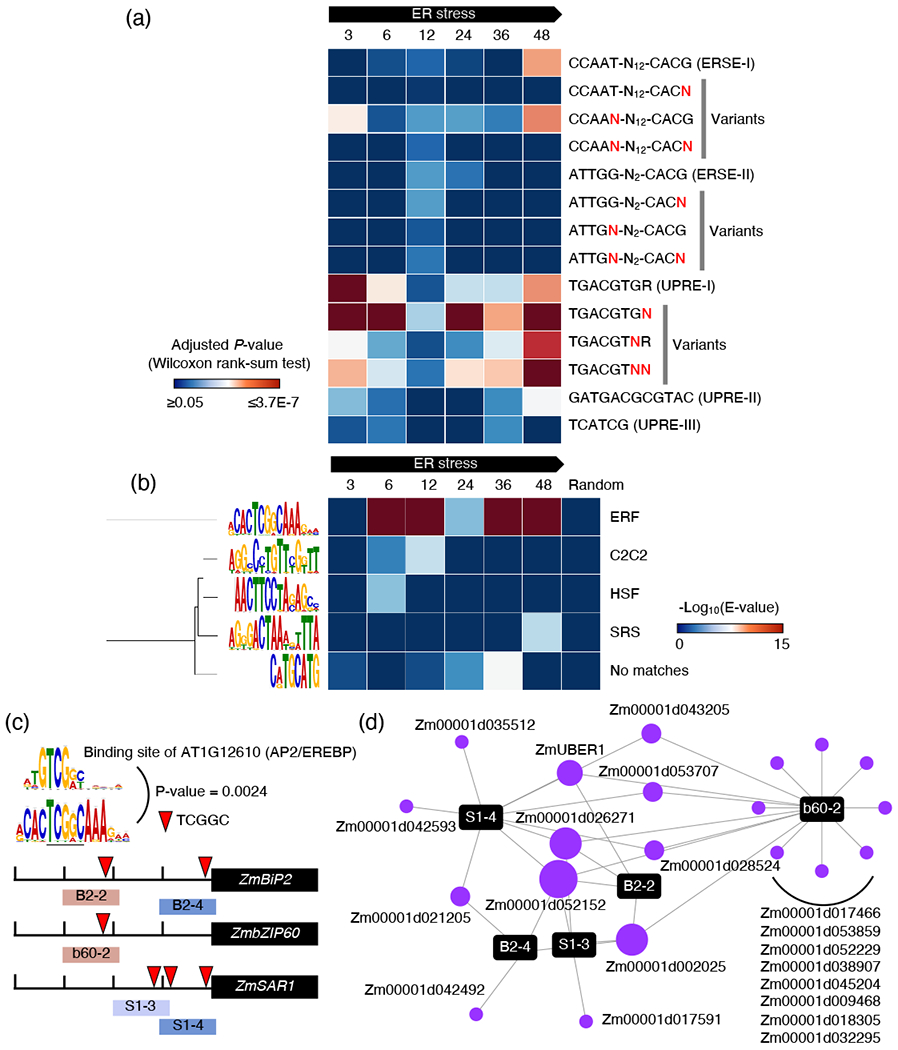
A TF network mediated by novel CRE underlying gene expression in the UPR. (a) Enrichments of canonical ER stress-responsive CRE with or without mismatch on 1-kb promoters of DEGs in maize (n = 1232 for 3 h, 2157 for 6 h, 351 for 12 h, 992 for 24 h, 1231 for 36 h and 2023 for 48 h). The analysis was carried out using AME (Buske et al., 2010) (Wilcoxon rank-sum test, Adjusted P-value by Bonferroni correction). N marked by red color indicates a random nucleotide. (b) Enrichments of de novo motifs on promoters of DEGs in maize during the adaptive ER stress. De novo motif analysis was performed using MEME (Bailey and Elkan, 1994). Motifs are displayed on the left of each row. The best match to a transcription factor family for the motif was identified from the Arabidopsis cistrome database (O’Malley et al., 2016) using TOMTOM (Gupta et al., 2007) and displayed to the right of each row. The median number of DEGs across time points is 280. We randomly selected 280 protein-coding genes in the maize genome and included the de novo motif analysis as a control. (c) Location of the highly enriched de novo motif on promoters of ZmBiP2, ZmbZIP60 and ZmSAR1. (d) A network of AP2/EREBP TFs on UPR promoters containing the de novo motif. The size of the nodes is proportional to the number of PDIs.
