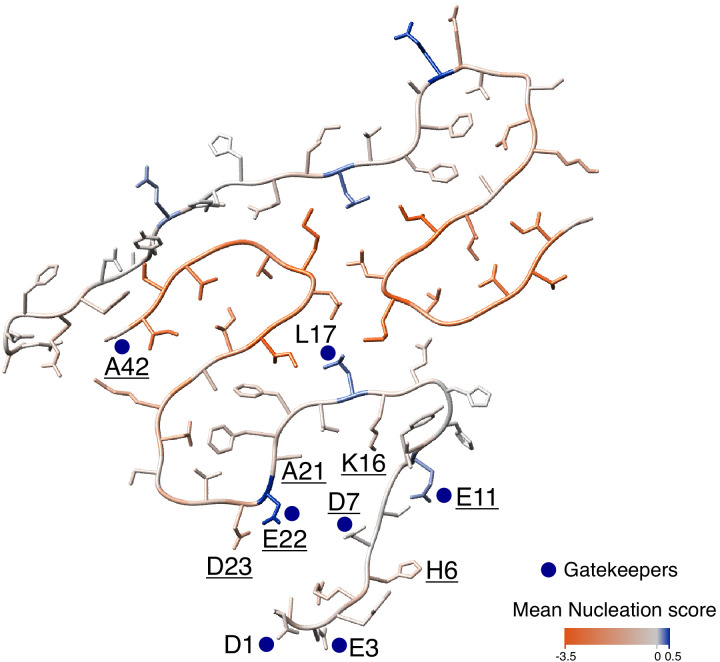
Figure 5. Mutational landscape of the amyloid beta (Aß) amyloid fibril.
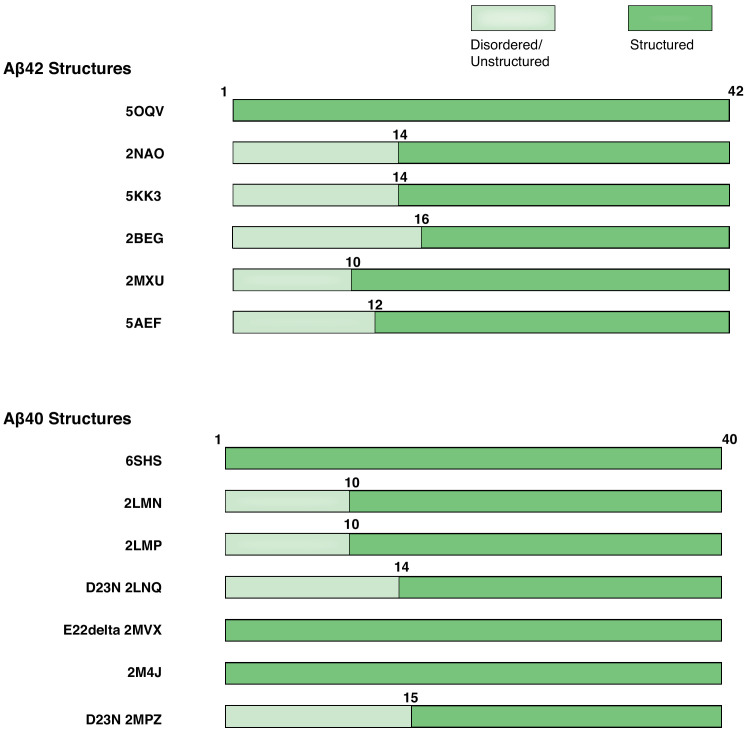
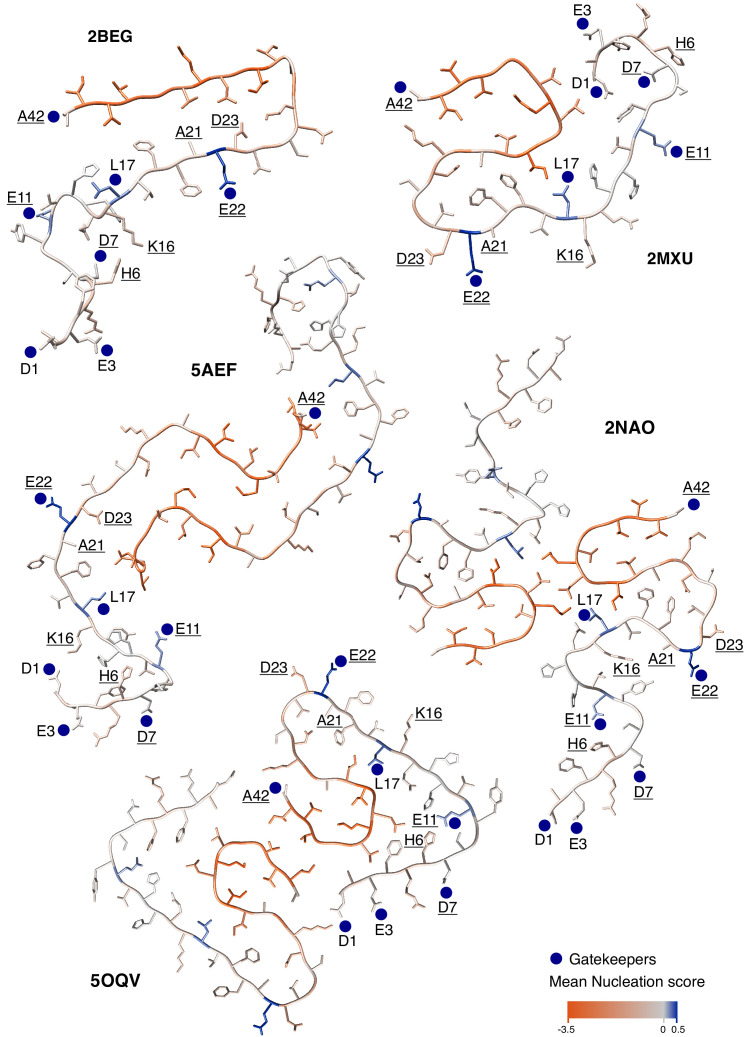
Average effect of mutations visualized on the cross-section of an Aß amyloid fibril (PDB accession 5KK3; Colvin et al., 2016). Nucleation gatekeeper residues and known familial Alzheimer’s disease (fAD) mutations positions are indicated by the wild-type (WT) aa identity on one of the two monomers; gatekeepers are indicated with blue dots and fAD are underlined. A single layer of the fibril is shown and the unstructured N-termini (aa 1-14) are shown with different random coil conformations for the two Aß monomers. See Figure 5—figure supplement 2 for alternative Aß42 amyloid polymorphs.