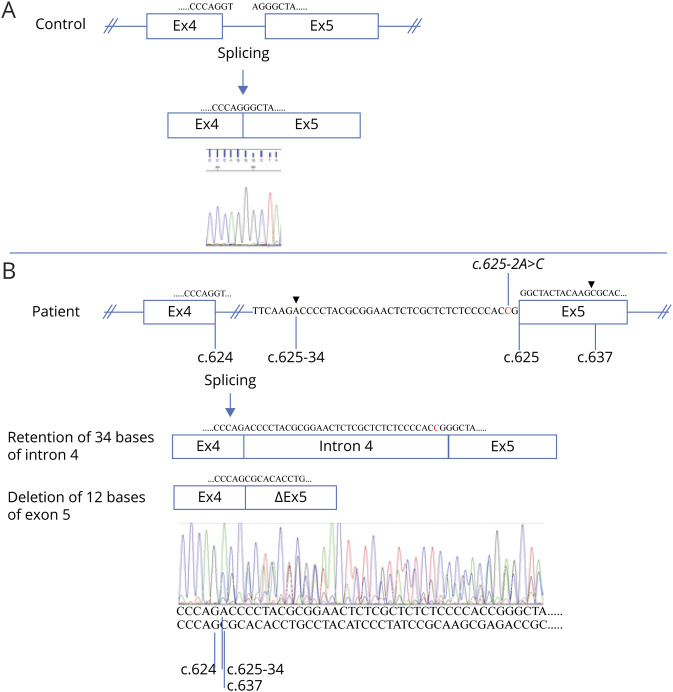
Figure 3. The c.625-2A>C Variant in COLGALT1 Results in 2 Aberrant Transcripts.
Reverse-transcriptase PCR and subsequent Sanger sequencing of Epstein-Barr virus–transformed cycloheximide-treated lymphocytes of a control (A) and the patient (B) resulted in normal splicing of intron 4 from the control and aberrant splicing for the patient. The 625-2A>C variant (indicated) resulted in the use of 2 cryptic splice sites (splicing positions indicated by arrow heads), 1 in intron 4, and 1 in exon 5. Thus, 2 populations of transcripts were detected in different quantities in the patient (compare peak heights). The major transcript contains 34 bases of intron 4 (retention) starting from position c.625-34 (indicated). The minor transcript lost 12 bases of exon 5, fusing c.624 to c.637 (indicated). Exon sizes are not to scale. Exon 4 (Ex4), intron 4 (indicated), exon 5 (Ex5), and exon 5 missing 12 bases (ΔEx5) are indicated by boxes, and partial sequences of these gene components are shown below the chromatograms and in the schematics.