Figure 1.
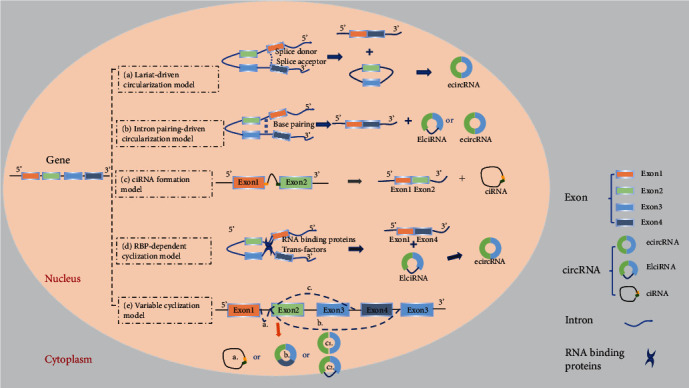
Schematic diagram illustrating five models of circRNA biogenesis. (a) Lariat-driven circularization model: the 3′ splice donor of exon 1 and the 5′ splice acceptor of exon 4 link up end-to-end by exon skipping and form an exon-containing lariat structure. Finally, the ecircRNA forms after introns are removed. (b) Intron pairing-driven circularization model: direct base pairing of introns forms a circulation structure, thereby forming ecircRNA or EIciRNA after intron removal. (c) ciRNA formation model: the elements near the splice site escape debranching stably so that the intron lariat is formed from the splicing reaction. (d) RNA binding protein- (RBP-) dependent cyclization model: RBPs bridge two flanking introns close together and then remove introns to form circRNAs. (e) Variable cyclization model: splicing selection and exon circularization may be influenced by inverted repeated Alu pairs (IRAlus) and the competition between them.
