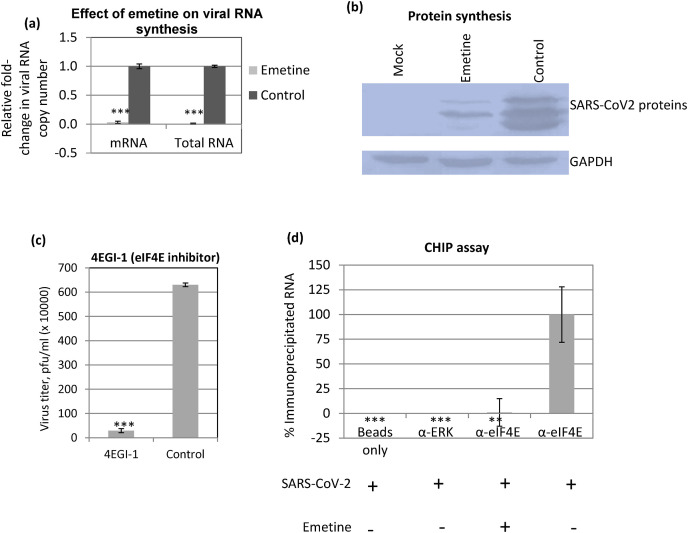
Fig. 2.
Emetine treatment results in decreased synthesis of SARS-CoV-2 RNA and proteins. (a) Effect of emetine on RNA synthesis: Confluent monolayers of Vero cells, in triplicates, were infected with SARS-CoV-2 for 1 h at MOI of 5. Emetine was added at 4 hpi and cells were harvested at 10 hpi to determine the levels of SARS-CoV-2 RNA by qRT-PCR. Threshold cycle (Ct) values were analysed to determine relative fold-change in copy numbers of total RNA and mRNA. Values are means ± SD and representative of the result of at least 3 independent experiments. Pair-wise statistical comparisons were performed using Student's t-test (*** = P < 0.001). (b) Protein synthesis: Vero cells were infected with SARS-CoV-2 at MOI of 5, followed by washing with PBS and addition of fresh MEM. Cell lysates were prepared at 12 hpi to detect the viral proteins by Western blot analysis by using serum derived from a COVID-19 positive patient. The serum reacted to produce at least three bands in the range of 40–60 kD (suggestive of SARS-CoV-2 “N” and/or “S” protein). The levels of viral proteins (upper panel), along with housekeeping GAPDH protein (lower panels) are shown. (c) eIF4E is required for efficient SARS-CoV-2 replication. Vero cells, in triplicates, were infected with SARS-CoV-2 at MOI of 0.1 in the presence of 0.5 μg/ml of 4EGI-1 (eIF4E inhibitor) or equivalent volume of DMSO (vehicle-control). The virus yield in the infected cell culture supernatant was quantified by plaque assay (*** = P < 0.001). (d) CHIP assay: Vero cells, in triplicates were infected with SARS-CoV-2 at MOI of 5 followed by washing with PBS and addition of fresh MEM containing emetine or vehicle control(s). At 10 hpi, cell lysates were prepared as per the procedure described for CHIP assay (materials and method section). The clarified cell lysates were incubated with α-eIF4E (reactive antibody), α-ERK (nonreactive antibody) or equivalent volume of IP buffer (Beads control) followed by incubation with Protein A Sepharose® slurry. The beads were then washed five times in IP buffer. To reverse the cross-linking, the complexes were then incubated with Proteinase K. Finally, the reaction mixtures were centrifuged and the supernatant was subjected to cDNA preparation and quantitation of SARS-CoV-2 RNA (E gene) by qRT-PCR. Values are means ± SD and representative of the result of at least 3 independent experiments. Pair-wise statistical comparisons were performed using Student's t-test ** = P < 0.01, *** = P < 0.001).