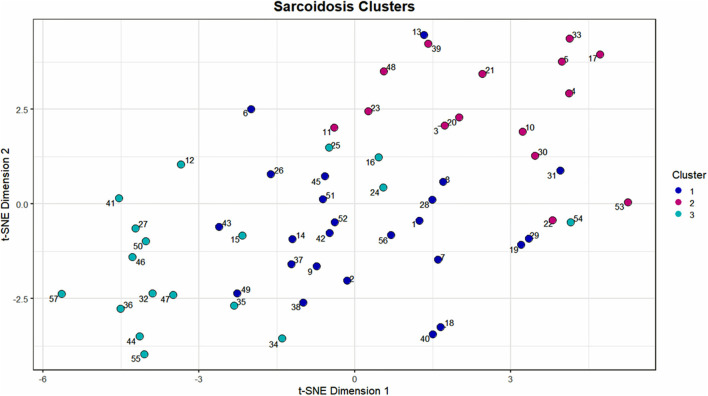
Figure 2.
Scatterplot visualization of the UIC-Sarcoidosis cohort over 2 dimensions utilizing the t-stochastic neighbor embedding (t-SNE) dimension reduction algorithm. The first and second dimensions are represented by the x-axis and y-axis, respectively. Points on the scatterplot represent individual subjects in the UIC-Sarcoidosis cohort and the distance between points is indicative of dissimilarity between subjects. Colors represent clusters identified by the Modha-Spangler algorithm utilizing partitioning around medoids with Gower's distance for base clustering of mixed data. Overall, Cluster 1 was identified as the largest cluster and comprised 41.38% of the cohort (24/58 subjects). Cluster 2 and 3 represented 25.86% (15/58) and 32.76% (19/58) of the cohort, respectively.