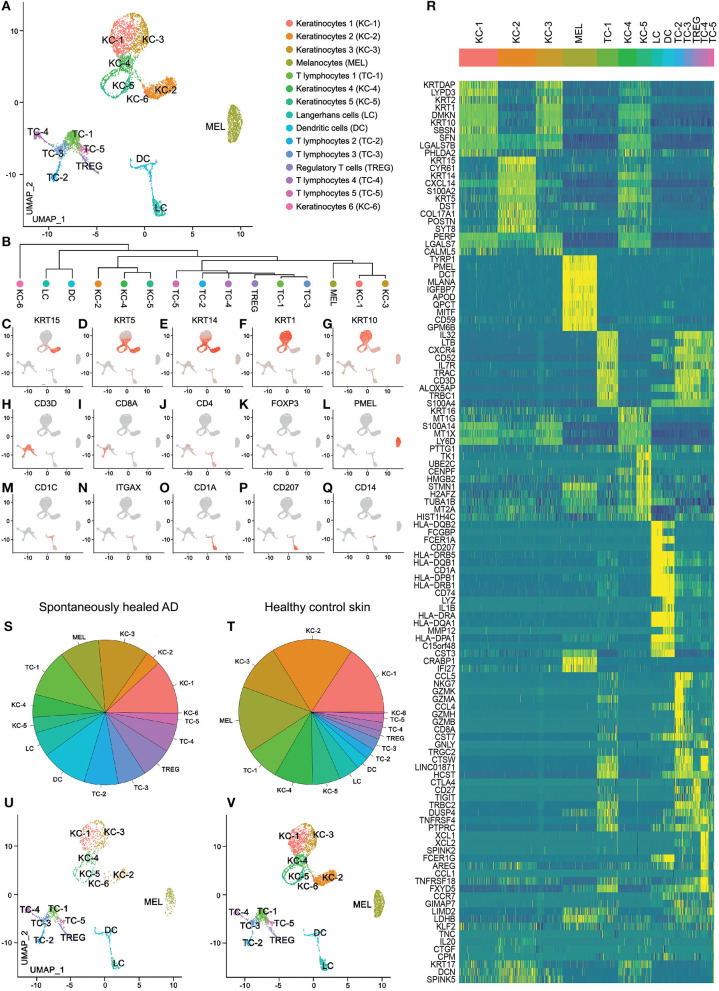
Figure 1.
scRNA-seq map of skin cells from suction blister samples of spontaneously healed atopic dermatitis and healthy control skin. (A) UMAP of 7,269 cells integrated from spontaneously healed atopic dermatitis patients (n = 4) and healthy control individuals (n = 4) according to similarity of their transcriptome, resulting in 15 different color-coded clusters. (B) Unsupervised hierarchical clustering showing relatedness of cell clusters (average gene signatures; correlation distance metric and average linkage). (C–Q) Feature plots combining all samples of expression distribution for cluster-specific marker genes. Intensity of expression levels for each cell is color-coded (red) and overlaid onto UMAP plots. (R) Heat map displaying the top 10 differentially expressed genes (according to highest log-fold change ordered by smallest adjusted p-value using logistic regression with Bonferroni correction for each cluster compared with the rest of the dataset); upregulation is indicated in yellow, and downregulation in blue; gene names are shown on the left. (S,T) Color-coded frequencies of cell clusters for spontaneously healed atopic dermatitis (n = 1,849) and healthy control blister samples (n = 5,420) displayed as pie graphs. (U,V) Separate UMAP plots of spontaneously healed atopic dermatitis and healthy control blister samples. AD, atopic dermatitis; KC, Keratinocytes; MEL, Melanocytes; TC, T lymphocytes; LC, Langerhans cells; DC, Dendritic cells; TREG, regulatory T lymphocytes; UMAP, Uniform Manifold Approximation and Projection.