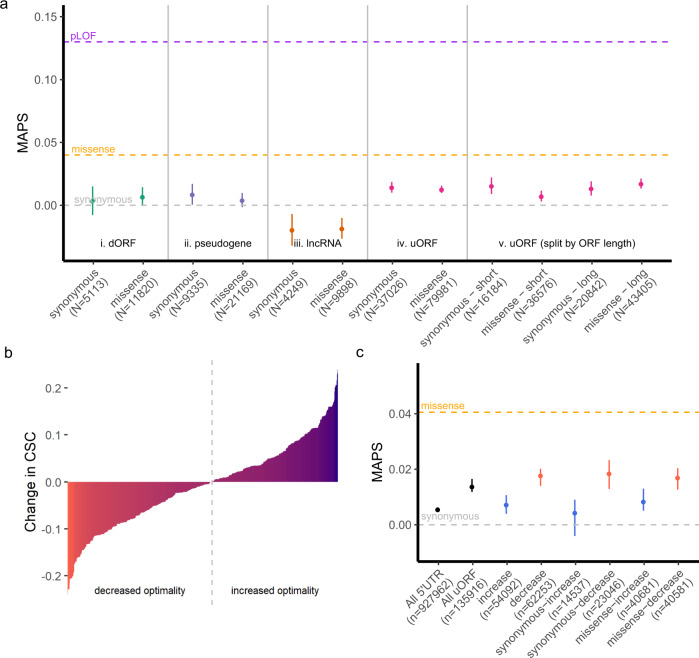
Fig. 2. uORFs do not exhibit strong selective pressure to maintain amino acid identity.
a Points representing MAPS scores for single nucleotide variants within each ncORF category separated by predicted consequence (synonymous or missense) in each ORF. (i–iv) Allele frequencies for predicted missense SNVs are not significantly enriched for singletons than those for predicted synonymous SNVs. (v) MAPS scores are no different for long uORFs (>118 codons) compared to the rest (short). Gray, orange, and purple dashed lines represent MAPS scores for synonymous, missense, and predicted loss-of-function (pLOF) SNVs affecting canonical protein coding sequences in gnomAD. Error bars represent bootstrapped 90% confidence intervals. b Translated uORF variants ranked by predicted change to codon optimality using codon stability coefficient (CSC) scores from SLAM-seq (red = decreasing, blue = increasing)34. Gray dotted line denotes boundary separating optimality increasing versus decreasing SNVs. c Points representing MAPS scores for SNVs separated by predicted consequence on codon optimality shows heightened constraint against decreasing optimality variants, while variants increasing optimality are indistinguishable from all 5′UTR variants. Error bars represent bootstrapped 90% confidence intervals.