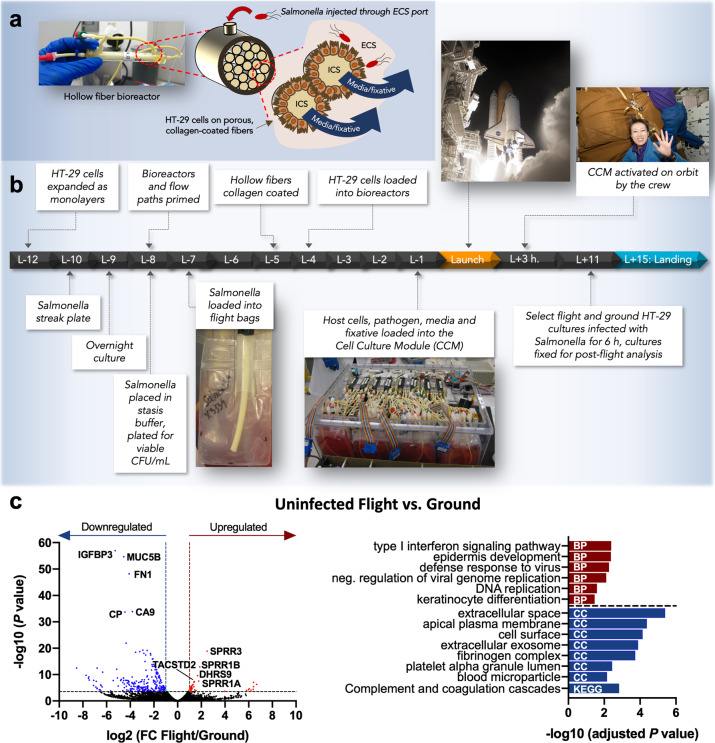
Fig. 1. STL-IMMUNE experimental design and impact of spaceflight on uninfected HT-29 transcriptional profiles.
a Hollow fiber bioreactor technology. Photo and cross-sectional schematic of the hollow fiber bioreactors used in this study. Hollow fibers had a pore size of 0.3 µm to retain bacteria within the extracapillary space (ECS) while still facilitating nutrient and gas exchange via diffusion as oxygenated media was perfused through the intracapillary space (ICS). b STL-IMMUNE experimental design. HT-29 cells and bacteria were prepared as shown (see “Methods” section for additional detail). Bioreactors, bacteria, and fixative bags were integrated into the CCM hardware and launched aboard Space Shuttle Discovery mission STS-131. Crew members activated the CCM once on orbit. At mission elapsed day 11, cultures were infected with S. Typhimurium for 6 h and fixed for postflight analysis. Launch and astronaut images: NASA [STS-131-S-036 (image is slightly cropped), S131E007206, and S131E007188]. c Spaceflight-induced transcriptional responses of HT-29 cells relative to ground controls. Left panel: volcano plot depicting differentially expressed genes (DEGs) between flight and ground control cultures (flight/ground). Red: significantly upregulated genes according to a log2 fold change in expression of at least ± 1 and FDR < 0.05. Blue: significantly downregulated genes. The top 5 significantly up and downregulated genes are labeled. Right panel: Analysis of upregulated (red) and downregulated (blue) transcripts revealed significantly enriched GO terms and KEGG pathways using DAVID. BP biological process, CC cellular component. The authors affirm that informed consent for publication of the images in Fig. 1 was provided.