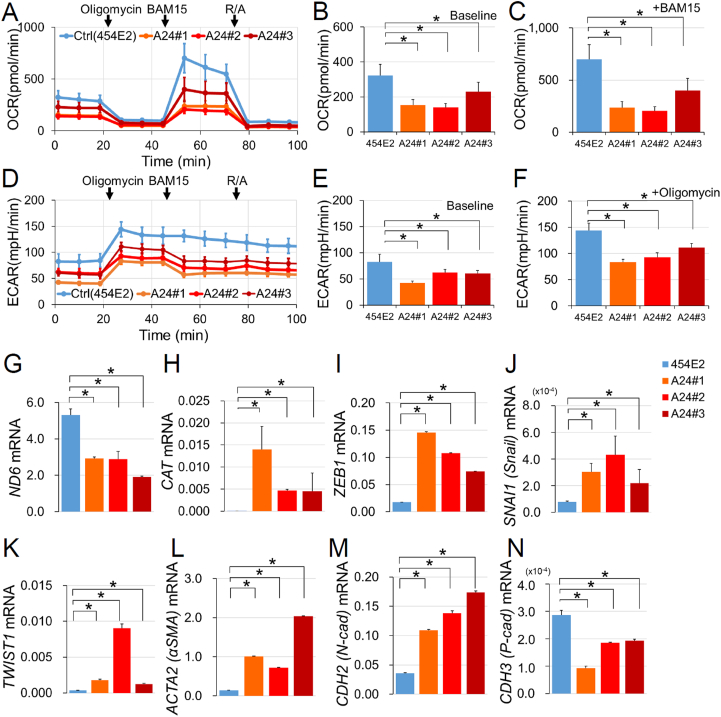
Fig. 5.
Respiratory deficit in the MELAS iPSC-derived RPE. (A–C) The oxygen consumption rate (OCR) and the (D–F) extracellular acidification rate (ECAR) in the control (454E2) and MELAS (A24#1–3) iPSC-derived RPE samples measured by an extracellular flux analyzer. Oligomycin (1 μM), BAM15 (10 μM), and R/A (rotenone, 2 μM; antimycin A, 2 μM) were sequentially added to the well. OCRs in the control (454E2) and MELAS (A24#1–3) iPSC-derived RPE were compared at baseline (B, at 1 min 23 s) and the maximal oxygen consumption condition after adding BAM15 (C, at 53 min 20 s). ECARs in the control (454E2) and MELAS (A24#1–3) iPSC-derived RPE were compared at baseline (E, at 1 min 23 s) and at the maximal extracellular acidification condition after the addition of oligomycin (F, at 27 min 21 s). n = 3–4. (G–N) mRNA levels of the mtDNA gene encoding NADH dehydrogenase 6 (ND6, G), the antioxidant enzyme catalase (CAT, H), as well as genes encoding epithelial-mesenchymal transition markers ZEB1 (I), SNAI1 (J), TWIST1 (K), ACTA (L), CDH2 (M), and CDH3 (N) in the control (454E2) and MELAS (A24#1–3) iPSC-derived RPE. n = 4. *P < 0.05.