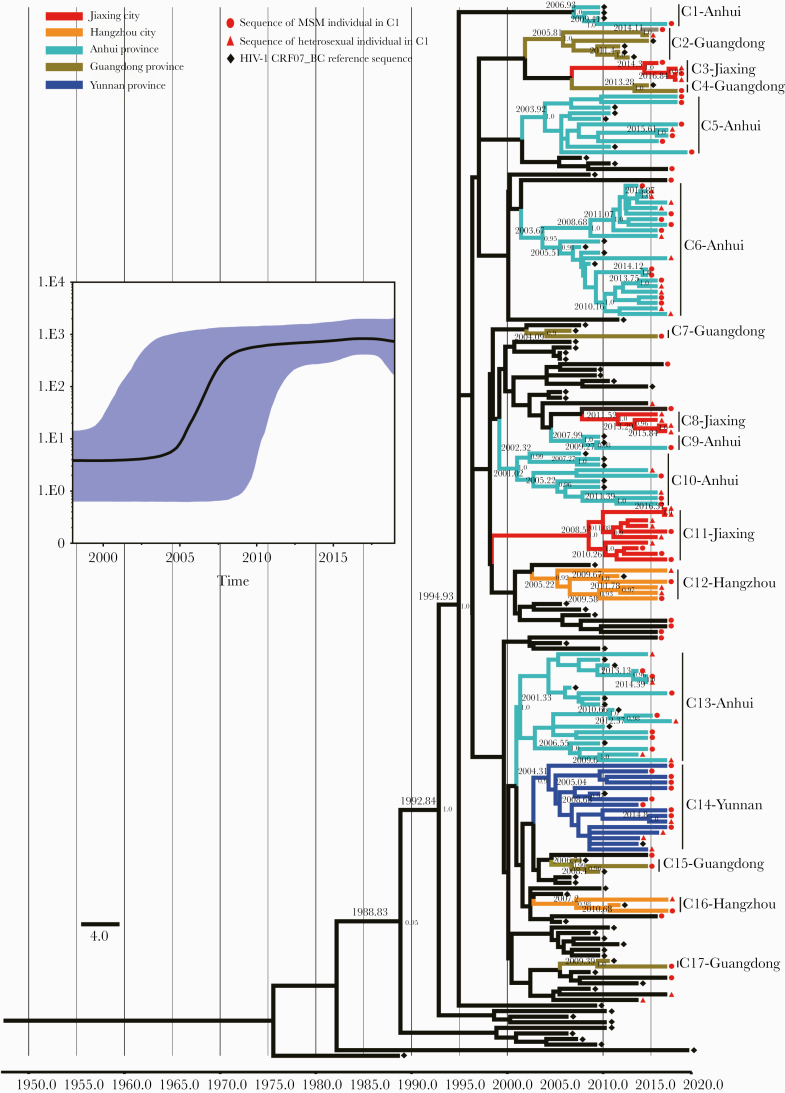
Figure 4.
Maximum clade credibility (MCC) tree of the active and large cluster C1 of CRF07_BC-infected patients based on HIV-1 pol sequences from 2015 to 2018. Bayesian analyses were performed using BEAST, version 1.75 (see the “Methods” for details). Bayesian skyline plot of the cluster C1 was demonstrated (the x-axis represents the time in units of years, and the y-axis represents the effective population size); the median effective population size is shown in black, and the 95% highest probability density (HPD) credible region is shown in blue. In the MCC tree, sequences from190 CRF07_BC sequences (85 reference sequences and 105 sequences of the cluster C1) form 17 unique phylogenetic clades, labeled C1 through C17; the branch lengths reflect evolutionary time, and nodes labeled with evolutionary time are supported by a high posterior probability (≥0.90). The corresponding time scale was marked at the bottom of the MCC tree; different colors of branches indicate that the formed clades contain reference sequences from different provinces/cities in China. Abbreviation: MSM, men who have sex with men.