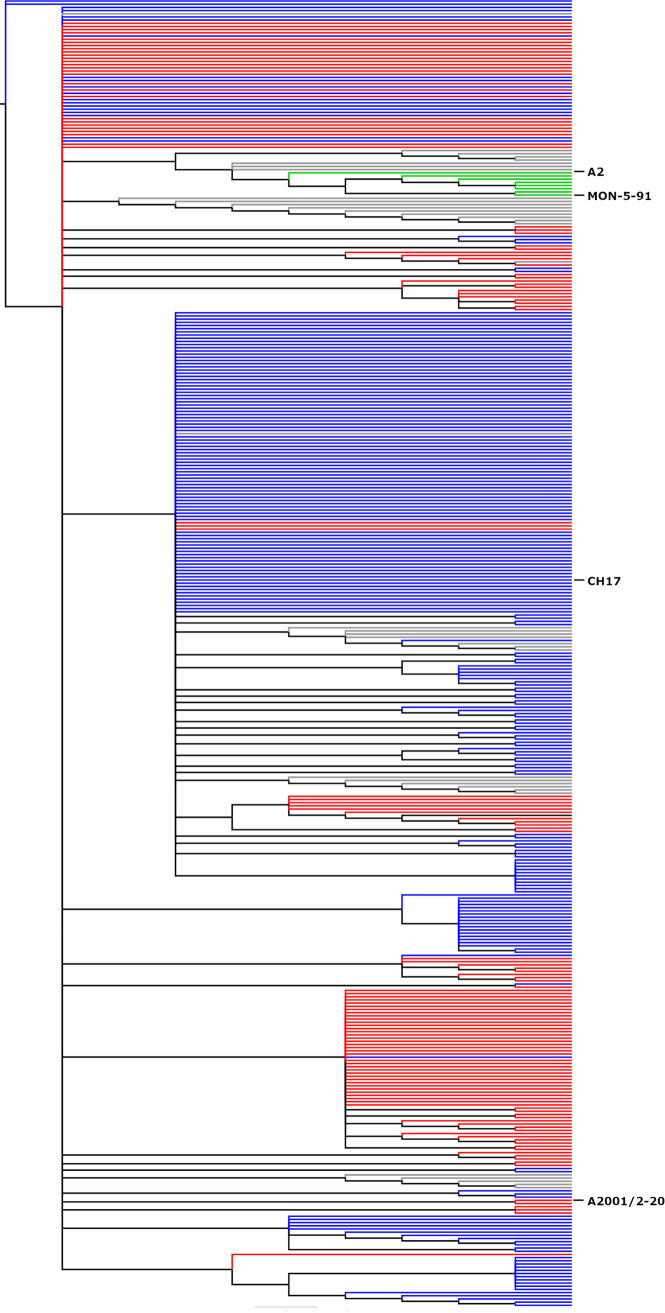
FIG 2.
Comparison of RSV A4G and 2stop+A4G genotype phylogenetics. The C-terminal 270 nucleotides of 411 RSV subgroup A isolates were aligned using ClustalW and phylogenetic reconstruction was performed using MEGA5 software by maximum likelihood (general time reversible model, 1,000 bootstrap replicates). The tree was constructed and annotated using FigTree v1.2.2. Gray nodes (9% prevalence [37/411]) represent strains, such as A2, containing A2-like G gene transcription and translation termination signals containing a single stop codon and an A nucleotide at the 4th position of the gene end transcription terminator. Blue nodes (56.6% prevalence [233/411]) represent A4G genotypes, including strain CH17, and red nodes represent 2stop+A4G genotypes (32% prevalence [132/411]), including strain A2001/2-20. Green nodes (2% prevalence [8/411]) represent wild-type strains, such as MON-5-91, that are distantly related to strains A2 and Long. Branch lengths are measured as number of substitutions per site (scale bar, 0.02).