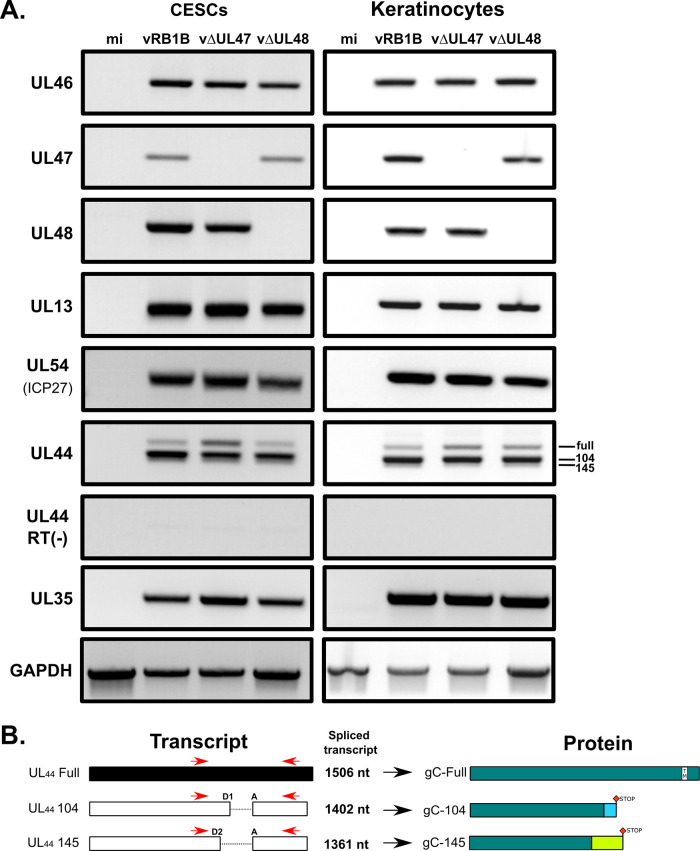
FIG 6.
Expression profiles of viral genes in cells infected with vRB1B, vΔUL47, or vΔUL48. (A) RNA was extracted from primary CESCs or chicken keratinocytes that were either mock infected or infected with vRB1B, vΔUL47, or vΔUL48. After cell lysis, RNA was extracted and used for RT-PCRs using primers to detect the UL46, UL47, UL48, UL44, UL13, and UL54 (ICP27). In addition, primers were used to detect cellular GADPH transcripts as a loading control and viral capsid protein gene UL35 as a control for infection levels. Reactions without reverse transcriptase [RT(–)] were also carried out to control for the absence of DNA contamination (here shown for UL44). (B) Splicing of the UL44 transcripts and associated predicted proteins. UL44 primers used for RT-PCR assays, as shown in panel A, are indicated in red. Two overlapping introns (104 and 145 bp) are present within the UL44 gene with a common acceptor site (A) and two donor sites (D1/D2). Splicing of these introns yields two transcripts of 1,402 and 1,361 bp, respectively, containing one frameshift each. The translation of these transcripts (UL44-104 and UL44-145) is expected to result in two truncated proteins of similar size but with differing C-terminal sequences (gC-104 and gC-145). The truncated proteins do not contain the transmembrane domain and are secreted.