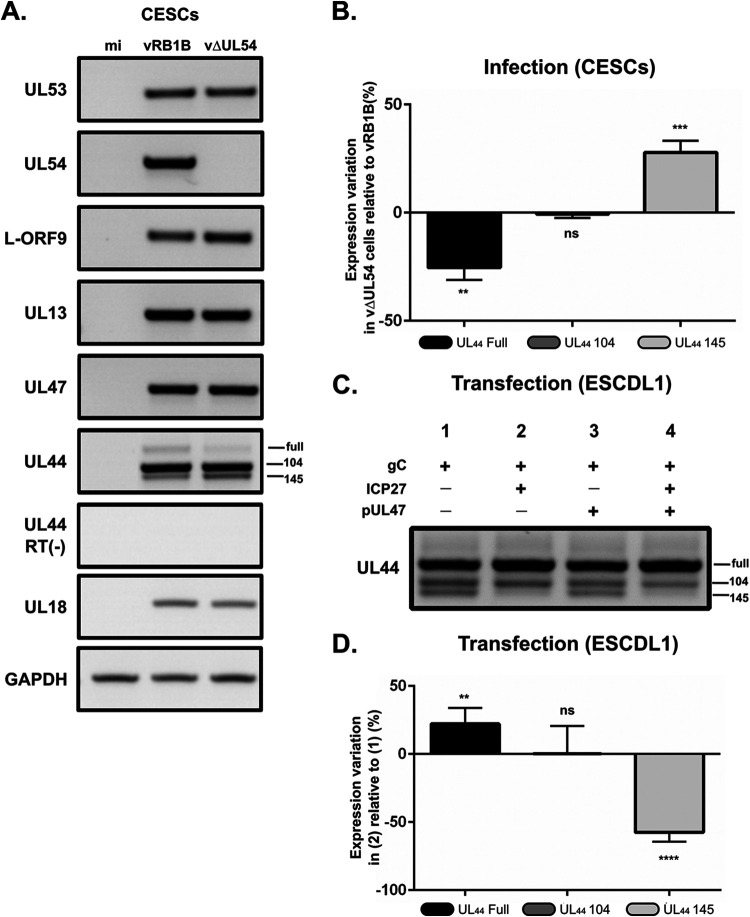
FIG 8.
ICP27 is another viral gene involved in horizontal transmission that affects splicing of UL44 transcripts. (A) RNA was extracted from primary CESCs that were either mock infected or infected with vRB1B or vΔUL54 and used for RT-PCRs using primers to detect the UL53, UL54, L-ORF9, UL13, UL47, and UL44 transcripts. In addition, primers were used to detect cellular GADPH transcripts as a loading control and viral capsid gene UL18 as a control for infection levels. Reactions without reverse transcriptase [RT(–)] were also carried out to control for the absence of DNA contamination (here shown for UL44). (B) RT-qPCR quantification of the UL44 transcripts using primers specific for each form. The abundance of each transcript form was normalized to the quantities of total UL44 transcripts. These normalized quantities in vΔUL54 were then quantified relative to the vRB1B levels (no variation = 0%). The experiment was done three times in triplicates. Error bars indicate standard deviations. Significance of differences was determined using a paired two-tailed Student t test on ΔCq values. **, P < 0.01; ***, P < 0.001; ns, not significant. (C) RNA was extracted from ESCDL-1 cells that were transfected with plasmids encoding gC and ICP27 and/or pUL47. RT-PCR was carried out using primers encompassing all three forms of UL44 transcripts as described in Fig. 6B. (D) The cDNA obtained under two conditions (gC alone) (condition 1) and (gC+ICP27) (condition 2) were used to quantify the different UL44 transcripts using primers specific for each form. The abundance of each transcript form was normalized to the quantities of total UL44 transcripts. These normalized quantities under condition 2 (gC+ICP27) were then quantified relative to the levels under condition 1 (gC alone) (no variation = 0%). The experiment was done three times in triplicates. Error bars indicate standard deviation. Significance of differences was determined by using a paired two-tailed Student t test on ΔCq values. **, P < 0.01; ****, P < 0.0001; ns, not significant.