Abstract
In addition to canonical TCR and BCR, cartilaginous fish assemble noncanonical TCR that employ various B-cell components. For example, shark T cells associate alpha (TCR-α) or delta (TCR-δ) constant (C) regions with Ig heavy chain (H) variable (V) segments or TCR-associated Ig-like V (TAILV) segments to form chimeric IgV-TCR, and combine TCRδC with both Ig-like and TCR-like V segments to form the doubly rearranging NAR-TCR. Activation-induced (cytidine) deaminase-catalyzed somatic hypermutation (SHM), typically used for B-cell affinity maturation, also is used by TCR-α during selection in the shark thymus presumably to salvage failing receptors. Here, we found that the use of SHM by nurse shark TCR varies depending on the particular V segment or C region used. First, SHM significantly alters alpha/delta V (TCRαδV) segments using TCR αC but not δC. Second, mutation to IgHV segments associated with TCR δC was reduced compared to mutation to TCR αδV associated with TCR αC. Mutation was present but limited in V segments of all other TCR chains including NAR-TCR. Unexpectedly, we found preferential rearrangement of the noncanonical IgHV-TCRδC over canonical TCR αδV-TCRδC receptors. The differential use of SHM may reveal how activation-induced (cytidine) deaminase targets V regions.
Keywords: Shark, Somatic hypermutation, T-cell receptor, TCRα/TCRδ locus, Thymus
Introduction
Jawed vertebrates evolved a sophisticated Ig superfamily (IgSF)-based adaptive immune system composed of B and T cells, a polymorphic and polygenic MHC, RAG-mediated somatic recombination, and activation-induced (cytidine) deaminase (AID)-mediated SHM [1–3]. This system relies on the rearrangement of variable (V), diversity (D), and joining (J) gene segments to generate the Ig heavy and light chains of BCR and the four canonical TCR chains during lymphocyte development [4–6]. Loci encoding each chain contain numerous V, (D), and J gene segments, and the resulting combinatorial potential results in a highly diverse immune repertoire (see Fig. 1) [7]. Each chain is encoded on separate loci (except TCR-δ, which is embedded within TCR-α) and loci are organized either as clusters of V, (D), and J segments followed by constant (C) region exons (V-D-J-C)n or as a contiguous translocon containing numerous V segments, (D segments), and J segments followed by C-region exons (VnDnJnC) [8, 9]. Lymphocytes further diversify antigen receptors during recombination by adding and subtracting nucleotides at gene segment joins, creating a unique third CDR3 that is highly variable in sequence and length. Traditionally, after gene recombination, Ig heavy chains (IgH) dimerize with Ig light chains (IgL) to form BCR expressed on the B-cell surface or antibodies secreted into the body humors, and TCR alpha (α) and beta (β) or gamma (γ) and delta (δ) chains dimerize to form canonical TCR expressed on the surface of T cells (see Fig. 1) [6, 10]. Together these mechanisms construct the efficient and effective adaptive immune repertoire necessary to respond to infection.
Figure 1.
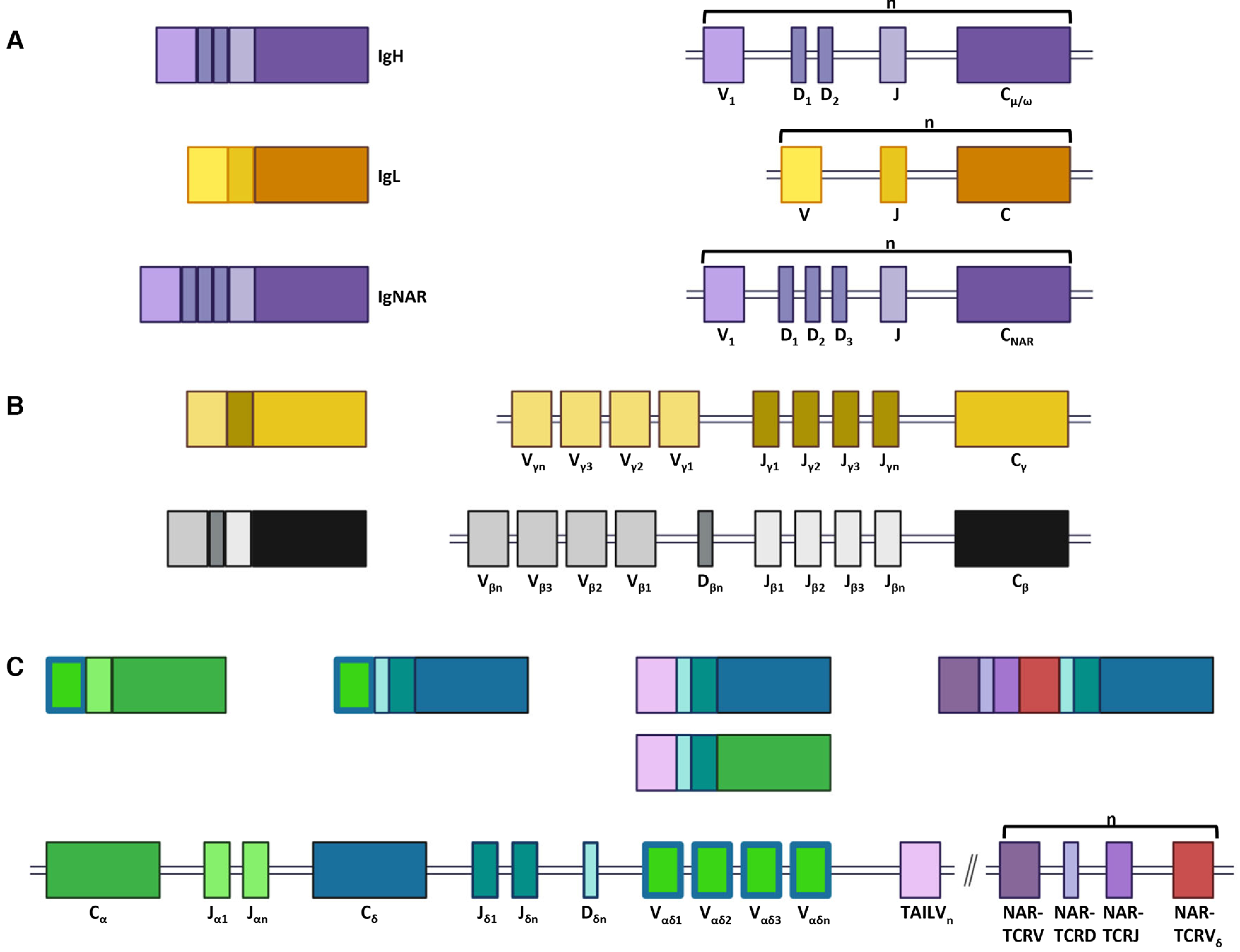
Nurse sharks generate complex B- and T-cell receptors. (A) Immunoglobulin (Ig) heavy (H, top) and light (L, middle) chains, and new (or nurse shark) antigen receptor (IgNAR, bottom) form from traditional B-cell components. B-cell (Ig) loci are organized in clusters of variable (V), (diversifying, D), and joining (J) gene segments followed by a constant region (VDJ-C or VJ-C, shown as a single cluster for each chain). (B) T-cell receptor (TCR) gamma (γ, top) and beta (β, bottom) chains form from traditional T-cell components located within individual translocons. (C) TCR delta (δ) chain is embedded within the TCR alpha (α) translocon and shares a common pool of V gene segments. Additionally, TCR-associated Ig-like V (TAILV) and both domains of NAR-TCR are found within this same locus. NAR-TCR gene segments occur in clusters of NARTCR V, NARTCR D, NARTCR J, and NAR-TCR Vδ (shown as a single cluster). Rearranged gene segments create traditional TCRα and TCRδ chains as well as unique, untraditional receptors (TAILVα or TAILVδ, and both domains of NAR-TCR). Further, IgHV-TCRδ rearrangements integrate components of IgH and TCR loci. VDJ gene segments and constant regions are color coded to correspond to the receptor chain they encode.
In B cells, receptor gene recombination occurs during lymphocyte development and cells exit bone marrow (or analogous primary lymphoid tissues, such as epigonal organ, in sharks) as näive lymphocytes with functional receptors. Exposure to antigen in the follicles of peripheral lymphoid tissue activates naïve mature B cells, stimulating BCR to undergo affinity maturation. During this process, AID catalyzes SHM of V regions followed by selection of the B cell, ultimately creating highly honed receptors for particular antigens [11]. Receptor gene recombination in T cells occurs similarly during thymic development. However, αβ TCR must undergo both positive and negative selection to ensure suitable binding to self-MHC but not to self-antigen; in this way, self-MHC referential yet self-tolerant T cells emerge from the thymus as mature cells [12, 13]. Research in mice and humans demonstrates that unsuccessful receptors can be rescued by further locus rearrangement (receptor editing), but ultimately most cells undergo apoptosis and are removed from the potential repertoire [14–17].
Recent studies in nurse sharks (Ginglymostoma cirratum) and other nonmodel vertebrates suggest that the boundaries between B- and T-cell components and repertoire diversification mechanisms are blurred in comparison to mouse and human. For example, marsupials and monotremes (e.g., Monodelphis domesticus, Ornithorhyncus anatinus) contain a unique TCRμ locus (TCRμ) that contains V, D, and J gene segments that somatically recombine, or are prejoined within germline DNA [18, 19], to form a receptor chain with two variable domains, the membrane-distal of which resembles IgH. Further, IgHV or Ig-like TCR-δ V segments (VHδ) are found in TCR-α/δ loci of all gnathostome groups except teleosts and placental mammals [20–26]. While many TCR-associating IgHV or VHδ genes are housed within the conventional αδ TCR locus, VHδ segments in Galliform birds are found in a second distinct TCR locus [22]. Nurse shark T cells assemble TCR using components traditionally considered BCR components, rearranging IgM or IgW (analogous to IgD) V segments to TCR alpha or delta constant (C) regions (see Fig. 1), though it remains unclear whether sharks are using IgHV only from within the conventional TCR-α/δ locus (cis-rearrangements) or are recombining Ig and TCR from separate loci (trans-rearrangements) as well [25, 26]. Doubly rearranging NAR-TCR, composed of a membrane-distal Ig-like NAR V domain and a proximal, supporting TCR δV domain, also marries unique Ig and TCR components into a single receptor (see Fig. 1) [27, 28]. Our lab recently discovered Ig-like V segments in nurse sharks that associate with TCR-α or -delta C regions (TCR-associated Ig-like V, TAIL V, see Fig. 1) [25]. Additionally, T cells can exploit BCR diversification mechanisms like AID-catalyzed SHM to generate additional thymic diversity. Chen et al. [29] presented definitive evidence that sandbar sharks utilize SHM to diversify gamma chain of γδ T cells, and camels employ SHM to diversify both TCR gamma and delta chains [29–33]. Additionally, nurse sharks utilize SHM for AID-catalyzed receptor salvaging to assist thymocytes through selection during thymic development [34]. Thus, gnathostome adaptive immunity displays remarkable elasticity in T-cell diversification mechanisms.
We examined a large dataset of TCR sequences to assess whether nurse sharks utilize SHM specifically for alpha-chain receptor salvaging or if SHM affects other canonical TCR chains and noncanonical receptors (IgH-TCRC rearrangements, NARTCR, and TAIL V-TCR C) alike. Additionally, this dataset compelled us to revise the current nomenclature for V gene segments within the alpha/delta (TCR-αδ) locus. Finally, we examine the use of SHM in light of the immunogenetic elasticity observed within the nurse shark TCR-αδ locus.
Results
Canonical nurse shark T cell receptor chains suggest few V segment families with many subfamilies
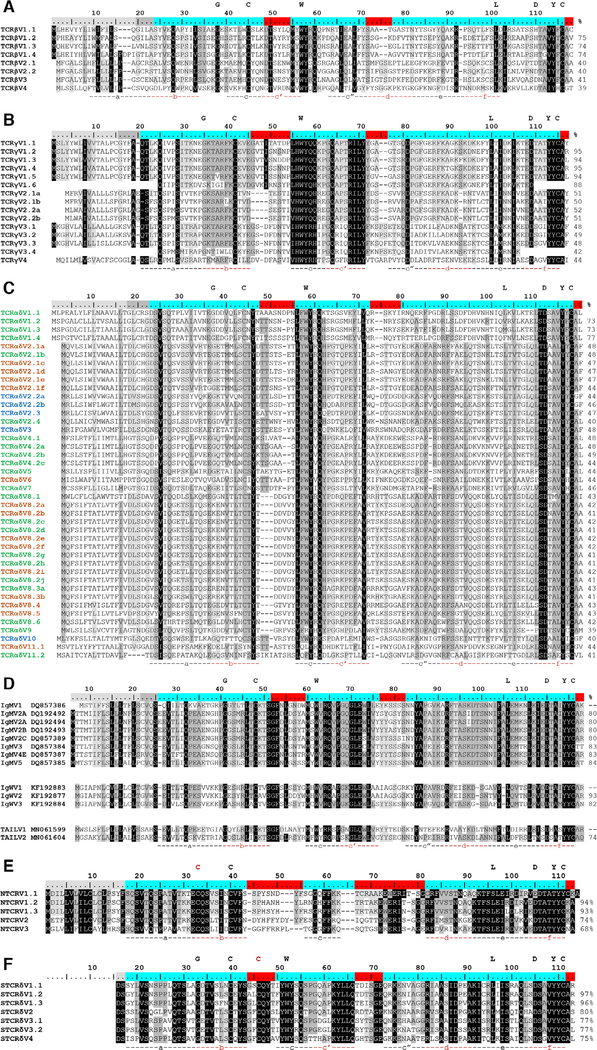
Our TCR data set contained 229 TCR-beta (TCR βV), 158 TCR-gamma (TCR γV), and 761 TCR-α/δ (TCR αδV) newly cloned or previously published V gene sequences (1149 total clones, see Supporting information Table 1 and 2). Using a refined approach to grouping V segments, we reduced the putative number of published TCR βV families to four, with TCR βV1 and TCR βV2 containing four and two subfamilies each, respectively (Fig. 2A; Supporting information Fig. S1A and S5). We reclassified TCR γV clones into four families (TCR γV1-TCR γV4) with multiple subfamilies in all but TCR γV4 (Fig. 2B; Supporting information Fig. S1B and S5). We did not identify any alleles for either chain. The 761 TCR-αδ clones sorted into 11 putative TCR αδV families and 24 subfamilies (Fig. 2C; Supporting information Fig. S2 and S5). Five of the 11 TCR αδV families spliced only to TCRα constant (C) region (TCR αδV 1, 4, 5, 7, and 9), two utilized only TCRδ C (TCR αδV 3 and 10), and four spliced interchangeably with TCR αC and TCR δC (TCR αδV 2, 6, 8, and 11). Five TCR αδV subfamilies included at least two alleles. For all canonical TCR chains, complete V gene segments contained the conserved tryptophan and two cysteine residues common to the IgSF domain (see Fig. 2) except TCR αδV11 (which includes only the conserved cysteine at position 104). This finding contrasts previous results indicating that TCR δV16 (TCR αδV8.2) lacks the first cysteine residue [26].
Figure 2.
Consensus sequence alignments for T-cell receptor V segments indicate substantial conservation between segments. V gene segments are grouped by identity for (A) beta (TCRβV) (B) gamma (TCRγV), (C) alpha/delta (TCRαδV), (D) immunoglobulin (Ig, IgMV and IgWV) and TCR-associated Ig-like V (TAILV), (E) NAR-TCR distal V domain (NTCRV), and (F) NAR-TCR proximal Vδ domain (STCRδV). V segment families share >70% nucleotide identity (e.g., TCRαδV2) and subfamilies have >80% nucleotide identity (e.g., TCRαδV2.1). Alleles share >90% nucleotide identity and common differences appear in more than one shark (e.g., TCRαδV2.1a). Letters above the scale denote conserved residues of antigen receptor domains. Regions below the alignment designate predicted beta strand location and direction. Shading within an alignment indicates amino acid conservation (Blosum62 score matrix [Threshold = 1]: black = 100% similar; dark grey = 80–100%; light grey = 60–80%). Values to the right of the alignments show the percent nucleotide identity to the first sequence. Highlighting within the scale indicates leader peptides (gray), framework regions (blue), and complementarity-determining regions (CDR, red). Coloring within the TCRαδV consensus sequence names identify the constant region used (green = TCR αC; blue = TCR δC; orange = both TCR αV and TCR δC). IgM or IgW germline sequence accession numbers are in sequence titles. IgM V2C is an Ig pseudogene due to defective Ig constant region exons but can form functional transcripts when associated with TCRα or TCRδ C. Each NAR-TCR V domain is encoded by V gene segments from a single gene family, but we employed original names to indicate the NARTCR cluster used. Gaps within a sequence are for alignment purposes only. Data from a single experiment, where each PCR tube represents a single replicate for each chain, shark, and tissue sample (see Supporting information Table 1 and 2).
Noncanonical T cell receptor variable gene segments are highly conserved
In addition to the canonical αβ and γδ TCR, we previously identified three “chimeric” nurse shark TCR containing Ig or Ig-like components: (1) IgHV can be associated with δC (or rarely, TCR-αC), rearranging an IgM or IgW (analogous to IgD) V segment to a TCRδC (or TCRαC) C region [26]; (2) doubly rearranging NAR-TCRs are composed of membrane-distal Ig-like V (NTCR V) and membrane-proximal or “supporting” TCR δV (STCR δV) [28]; and (3) TAIL V segments recombine Ig-like V and D segments to TCR J segments and can associate with either TCR αC or TCR δC regions (see Fig. 1) [25]. We sequenced 195 IgMV-TCR δC, 77 IgWV-TCR δC, 69 NAR-TCR (51 NTCR V and 62 STCR δV complete domains), and nine TAIL V clones (Supporting information Table S1 and S2).
IgHV-TCR δC clones aligned with five of the six canonical IgM germline groups (IgM V1-V5) and three of the six canonical IgW groups (IgW V1-V3) [35]. We identified three IgM V2 subfamilies and two IgW V1 subfamilies in our dataset (Fig. 2D; Supporting information Fig. S3 and S5). IgM V2C is an Ig pseudogene (due to defective Ig constant region exons), but we observed functional transcripts associated with TCR δC. Interestingly, our 5’ RACE libraries primed with TCR δC-specific primers generated more clones associated with IgM/IgW V segments (58%) than to canonical TCR αδV segments. These libraries comprised data from two “young” sharks (Tom Thumb, a neonate and Florence Nightingale, 3’ in length) and two “old” sharks (White and Grumpy, both greater than 8’ in length). Libraries from younger sharks generated more canonical TCR αδV-TCR δC arrangements (69 of 115 clones, 60%) and those from older sharks generated more noncanonical IgHV-TCR δC arrangements (75 of 126 clones, 60%). However, further study characterizing IgHV-TCR δC rearrangements is required to verify these observations.
Based on our conservative naming strategy, all NTCR V gene segments belonged to a single family containing three subfamilies, and subfamily NTCR V1 included four different alleles (Fig. 2E; Supporting information Fig. S4 and S5). All “supporting” V gene segments (STCR δV) comprised a single gene family composed of four subfamilies. Both STCR δV1 and STCR δV3 contained multiple alleles. However, we retained subfamily names in V segment identities for consistency with published data. We observed multiple combinations between NTCR V and STCR δV domains, but in general NTCR V1 associated with STCR δV1 (NTCR V1.1- STCR δV1.1a, NTCR V1-STCR δV1.1b; NTCR V1.2-STCR δV1.1b; NTCR V1.3-STCR δV1.2; NTCR V1.4-STCR δV1.3), NTCR V2 associated with both STCR δV2 and STCR δV4, and NTCR V3 associated with STCR δV3 (V3.1 and V3.2). In addition to the conserved tryptophan and two cysteine residues found in other TCR, all functional NAR-TCR sequences contained the noncanonical interdomain cysteine in FR1 of NTCR V (and CDR1 of STCR δV) required for domain stability [36].
Hotspot motifs in nurse shark T cell receptor variable segments do not necessarily predict mutation
AID preferentially alters C and G residues of WRCH/DGYW motifs of antigen receptors [37]. The number of WRCH/DGYW AID hotspot motifs in CDR did not differ from FR motifs in any of the canonical TCR V segments (see Fig. 3, Table 1). However, FR2 of TCR βV contained more WRCH/DGYW motifs than other FR. As expected, both CDR of IgHV contained more motifs than FR domains. NAR-TCR domains (NTCR V and STCR δV) contained the fewest WRCH/DGYW motifs in any region of all V segment types and within NTCR V, most WRCH motifs overlapped DGYW motifs, a pattern not observed in other V gene segments. Motif patterns did not vary by region. Thus, motif patterns alone do not predict mutation.
Figure 3.
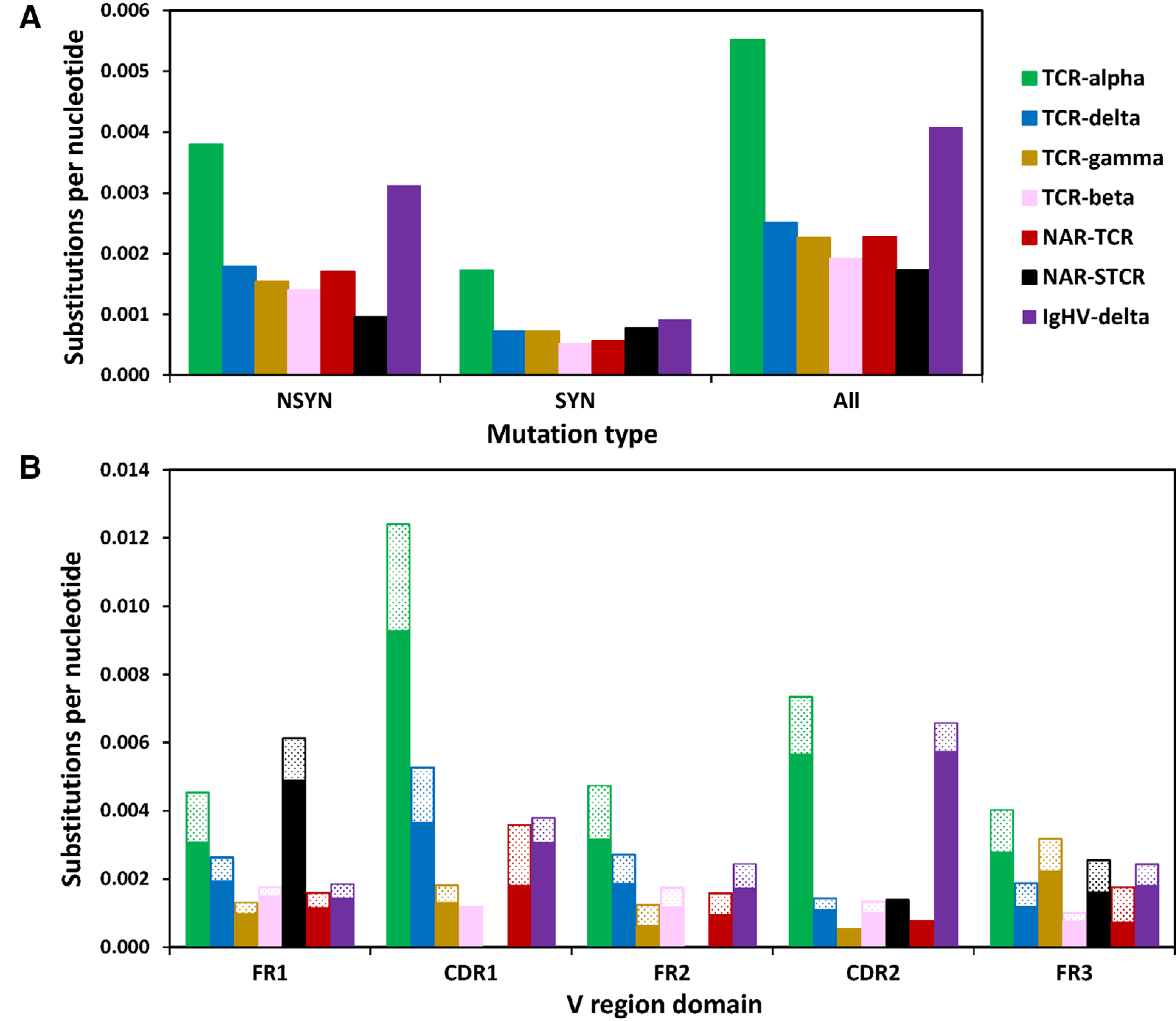
SHM targets complementarity-determining regions (CDR) of TCR-α associated with alpha constant regions. (A) TCR-α accumulates significantly more nonsynonymous (NSYN, solid) mutations than all other TCR chains except IgHV-δ sequences and significantly more synonymous (SYN, stippled) mutations than all other chains. (B) TCR-α/δ V gene segments associated with TCR-α constant (C) regions accumulate significantly more mutations in both framework (FR) and CDR than when associated with TCR-δ C, and in general, accumulate significantly more mutation than all other TCR chains except CDR2 domains of IgHV-δ (Student’s one-way, unpaired t-test, p < 0.01). We counted the number of mutations within 422 TCR-α/δ V-TCR-α C (green), 137 TCR-α/δ V-TCR-δ C (blue), 158 TCR-γ V (gold), 237 TCR-beta V (pink), 51 NARTCR V (red), 62 supporting NARTCR-δ V (NAR-STCR, black), and 275 IgHV-TCR-δ C (purple) sequences (Student’s one-way, unpaired t-test, *p < 0.05; **p < 0.01). Data from a single experiment, where each PCR tube represents a single replicate for each chain, shark, and tissue sample (see Supporting information Tables 1 and 2).
Table 1.
Target nucleotide mutation frequency in DGYW/WRCH mutation hotspots within framework regions (FR) and complementarity-determining regions (CDR) of T cell receptor (TCR) variable region (V) segments
| V segment | Hotspot motif | Region | # G/C Bases | % G/C | S/N | Observed G/C mutations | Expected G/C mutations | MIa) | χ2 pb) |
|---|---|---|---|---|---|---|---|---|---|
| TCRαδV-TCRαC | DGYW/WRCH | FR | 11008 | 1.9 | 1.27 | 140 | 5.9 | 23.62 | 0.0000 |
| CDR | 2997 | 0.5 | 1.94 | 58 | 2.6 | 22.73 | |||
| All | 14005 | 2.4 | 1.41 | 198 | 8.3 | 23.96 | 0.0000 | ||
| Outside motif | 568931 | 0.03 | 146 | 335.7 | 0.43 | ||||
| TCRαδV-TCRδC | DGYW/WRCH | FR | 4012 | 4.2 | 0.65 | 26 | 2.3 | 11.15 | 0.0000 |
| CDR | 714 | 0.7 | 0.84 | 6 | 0.5 | 11.15 | |||
| All | 4726 | 4.9 | 0.68 | 32 | 2.9 | 11.15 | 0.0000 | ||
| Outside motif | 90762 | 0.03 | 26 | 55.1 | 0.47 | ||||
| TCRβV | DGYW/WRCH | FR | 5476 | 9.8 | 0.29 | 16 | 5.5 | 2.92 | 0.0000 |
| CDR | 925 | 1.7 | 0.32 | 3 | 0.7 | 4.28 | |||
| All | 6401 | 11.5 | 0.30 | 19 | 6.2 | 3.07 | 0.0000 | ||
| Outside motif | 49435 | 0.07 | 35 | 47.8 | 0.73 | ||||
| TCRγV | DGYW/WRCH | FR | 3099 | 5.6 | 0.42 | 13 | 3.4 | 3.78 | 0.0000 |
| CDR | 581 | 1.0 | 0.69 | 4 | 0.4 | 11.27 | |||
| All | 3680 | 6.6 | 0.46 | 17 | 3.7 | 4.58 | 0.0000 | ||
| Outside motif | 51898 | 0.08 | 39 | 52.3 | 0.75 | ||||
| IgHV-TCRδC | DGYW/WRCH | FR | 4604 | 6.2 | 0.56 | 26 | 6.0 | 4.37 | 0.0000 |
| CDR | 1995 | 2.7 | 1.10 | 22 | 3.5 | 6.33 | |||
| All | 6599 | 8.9 | 0.73 | 48 | 9.4 | 5.12 | 0.0000 | ||
| Outside motif | 67254 | 0.08 | 57 | 95.6 | 0.60 | ||||
| NTCRV | DGYW/WRCH | FR | 644 | 14.6 | 0.47 | 3 | 1.7 | 1.80 | 0.2240 |
| CDR | 102 | 2.3 | 2.94 | 3 | 2.1 | 1.44 | |||
| All | 746 | 16.9 | 0.80 | 6 | 3.6 | 1.69 | 0.1542 | ||
| Outside motif | 3664 | 0.41 | 15 | 17.4 | 0.86 | ||||
| STCRδV | DGYW/WRCH | FR | 1019 | 11.3 | 0.20 | 2 | 1.4 | 1.40 | 0.5413 |
| CDR | 279 | 3.1 | 0.00 | 0 | 0.1 | 0.00 | |||
| All | 1298 | 14.3 | 0.15 | 2 | 1.6 | 1.27 | 0.7161 | ||
| Outside motif | 7754 | 0.12 | 9 | 9.4 | 0.96 |
DGYW/WRCH (G/C is the mutable position; D = A/G/T, Y = C/T, W = A/T, R = A/G, and H = T/C/A); “ALL” refers to G and C nucleotides found within hotspot motifs along the entire V segment (FR and CDR); “Outside motif” refers to G and C nucleotides outside a hotspot motif; S/N = substitutions per nucleotide; MI = mutability indexa); %G/C = proportion of all nucleotides in that category that are G or C. TCRαδV-TCRαC, alpha; TCRαδV-TCRδC, delta; TCRβV, beta; TCRγV, gamma; IgHV-TCRδC, trans-rearrangements between immunoglobulin heavy chain (IgH) and delta TCR constant regions; NTCRV and STCRδV, components of NARTCR.
Mutability index is the observed number of mutations of a specific nucleotide divided by the expected number of mutations of that nucleotide, with a value of 1.00 indicating random mutation.
χ2 analysis was used to compare observed and expected numbers of mutations between (a) FR and CDR regions and (b) all mutations inside and outside hotspot motifs for each V segment type.
Mutation occurs in TCR αδV associated with TCR αC but not with TCR δC or other canonical TCR chains
SHM within TCR was first identified in TCRγ chain of sandbar sharks [29, 30]. However, although mutation appeared to target nucleotide motifs preferred by AID (WRCH/DGYW), mutation tended not to result in aa replacement within CDR, a requisite for paratope changes during affinity maturation. Rather, sandbar sharks appeared to use SHM to generate a more diverse repertoire [29, 30]. We previously confirmed that SHM occurred within TCRγ and TCR δV segments of nurse shark, but we found that SHM altered TCR αV far more than it did gamma or delta, with replacement mutation targeting AID-preferred motifs of CDR within the thymus. This suggested that TCR-α likely uses SHM to salvage failing receptors during thymic selection [34]. In both BCRs and TCRs of sharks, SHM can occur as single point mutations or tandem mutations of two or more contiguous nucleotides, indicating at least two different cellular mechanisms generate mutations [38–40].
We first attempted to corroborate earlier findings of SHM in canonical TCR of nurse sharks. However, despite having unique CDR3 regions, TCRβ and TCRγ showed very little variation within V segment nucleotide sequences (see Supporting information Table S1), with about 0.002 substitutions per nucleotide (S/N) for both chains (see Fig. 3 and 4, Supporting information Fig. S6A). While we observed contiguous mutations within both chains, the majority of mutation occurred as single base changes and most base changes resulted in aa replacement (R) rather than silent (S) mutation (TCR βV: R/S = 2.7; TCR γV: R/S = 2.1; Supporting information Fig. S6A). As previously observed, TCR γV segments contained substantially more mutation than TCR βV segments. While TCR βV sequences accumulated more mutations to FR1 (0.0018 S/N) and fewer to CDR1 (0.0012 S/N; Fig. 3 and 4), TCR γV mutation was highest in FR3 (0.0032) and lowest in CDR2 (0.0005; Fig. 3 and 4). Most mutations in TCR βV (61%) and TCR γV (67%) were to C and G nucleotides, though only 35% of C/G mutations in TCR βV and 30% in TCR γV actually occurred within WRCH/DGYW motifs (Fig. 3 and 4; Table 2). Both TCR βV and TCR γV mutations were biased toward transitions (TCR βV FR: 49%, CDR: 44%; TCR γV FR:56%, CDR:63%; Table 2). TCR βV4 sequences exhibited the most mutation, with 64% of sequences (23 of 36) containing at least one nucleotide change (Supporting information Fig. S1). Again, we observed no mutation within any TCR γV4 segment (except a single base change in a leader region), suggesting this V segment may be useful as a partner chain with noncanonical receptors or with receptors highly specific for particular antigen.
Figure 4.
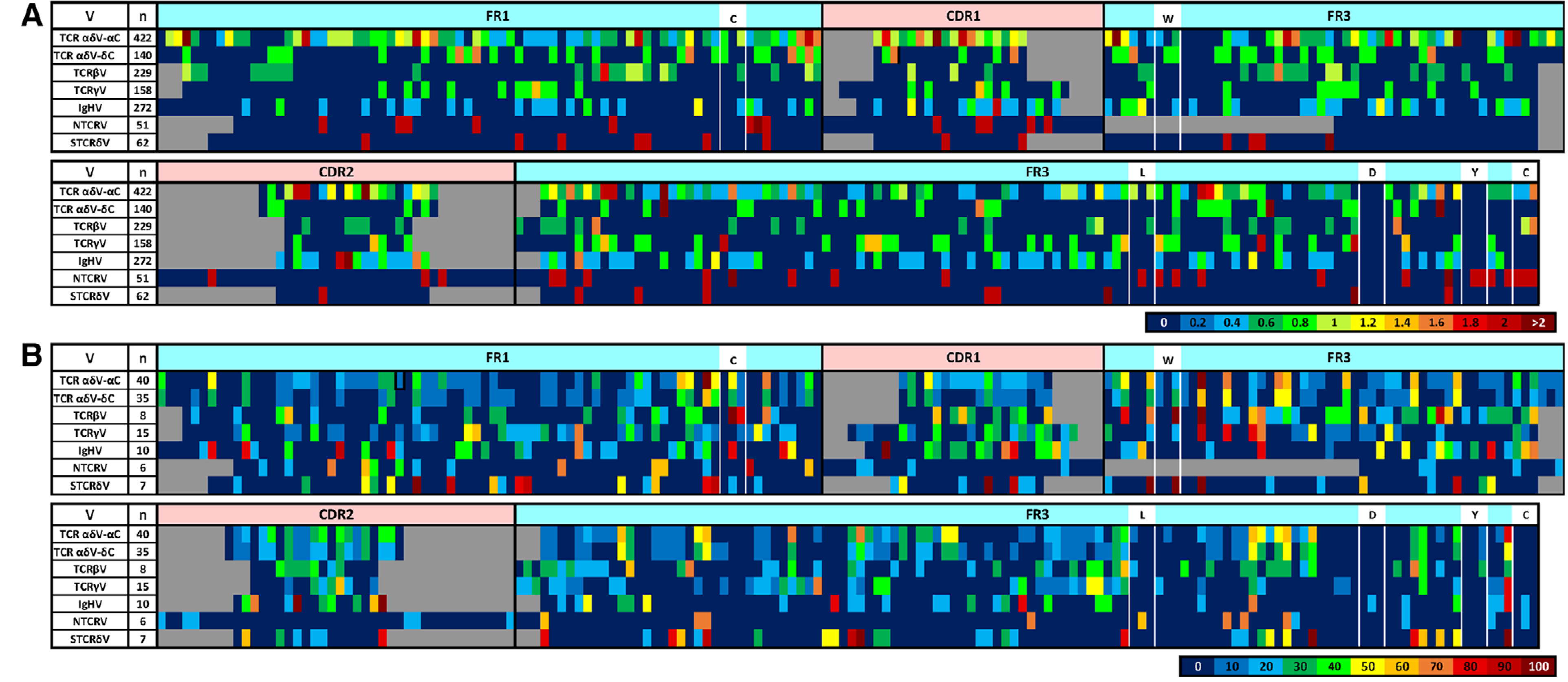
Heatmap coloring indicates (A) the proportion of sequences (n = number of sequences represented) within each segment type that contain a mutation or the (B) proportion of Vs within each segment type (n = number of V family groups) that contain a G/C target (within a DGYW/WRCH motif) at any single position along the sequence length (5’ to 3’). White boxes indicate the locations of conserved antigen receptor domain residues. Putative locations of framework regions (FR, light blue) and complementarity-determining regions (CDR, pink) are indicated above the heatmaps. Gaps (grey space) within the sequences are for alignment purposes only (segments are aligned by domain and conserved residue placement only). Numbers within each color scale represent the greatest value for that color. TCR, T-cell receptor, V, V segment, TCR αδV, TCR-α/δ V; TCR βV, TCR-beta V, TCR γV, TCR-gamma V; IgHV, immunoglobulin heavy chain V; NTCR V, NAR-TCR V; STCR δV, supporting TCR-δ V (NTCR V-STCR δV for doubly rearranged NAR-TCR). Data from a single experiment, where each PCR tube represents a single replicate for each chain, shark, and tissue sample (see Supporting information Table 1 and 2).
Table 2.
Mutation bias within T-cell receptor variable (TCRV) segments differed by V segment type
| V segment | Region | % Transition mutations | % Transversion mutations | % All mutations to G or C nucleotides | % G and C mutations inside motifs |
|---|---|---|---|---|---|
| TCRαδV-TCRαC | FR | 45.58 | 43.54 | 59.41 | 53.44 |
| CDR | 43.40 | 43.40 | 51.57 | 70.73 | |
| All | 45.00 | 43.50 | 57.33 | 57.56 | |
| TCRαδV-TCRδC | FR | 53.33 | 34.67 | 62.67 | 55.32 |
| CDR | 52.94 | 47.06 | 64.71 | 54.55 | |
| All | 53.26 | 36.96 | 63.04 | 55.17 | |
| TCRβV | FR | 48.75 | 36.25 | 60.00 | 33.33 |
| CDR | 44.44 | 44.44 | 66.67 | 50.00 | |
| All | 48.31 | 37.08 | 60.67 | 35.19 | |
| TCRγV | FR | 56.00 | 34.67 | 65.33 | 26.53 |
| CDR | 62.50 | 25.00 | 87.50 | 57.14 | |
| All | 56.63 | 33.73 | 67.47 | 30.36 | |
| IgHV-TCRδC | FR | 50.72 | 37.68 | 49.28 | 38.24 |
| CDR | 51.92 | 46.15 | 71.15 | 59.46 | |
| All | 51.05 | 40.00 | 55.26 | 45.71 | |
| NTCRV | FR | 42.86 | 47.62 | 42.86 | 30.00 |
| CDR | 62.50 | 25.00 | 37.50 | 27.27 | |
| All | 48.28 | 41.38 | 41.38 | 28.57 | |
| STCRδV | FR | 40.91 | 54.55 | 45.45 | 20.00 |
| CDR | 66.67 | 33.33 | 33.33 | 0.00 | |
| All | 44.00 | 52.00 | 44.00 | 18.18 |
FR, framework region; CDR, complementarity-determining region; TCRαδV-TCRαC, alpha; TCRαδV-TCRδC, delta; TCRβV, beta; TCRγV, gamma; IgHV-TCRδC, trans-rearrangements between immunoglobulin heavy chain (IgH) and TCRδ constant regions; NTCRV and STCRδV, distal and proximal (supporting) components of NAR-TCR.
We analyzed mutation data for V segments within the TCRαδ locus only if we could identify with certainty the constant region associated with the V segment. Thus, we included 422 TCR αδV associated with TCR αC and 140 TCR αδV associated with TCR δC in our analyses. We first confirmed previous results that CDRs of TCR αδV-TCR αC mutated significantly more than FRs (Fig. 3 and 4), with CDR1 accumulating the most mutation [34]. Within the TCRαδ locus, TCR αδV-TCR αC accrued more than twice as many mutations (0.0055 S/N) as TCR αδV-TCR δC (0.0025; p = 0.002; Fig. 3 and 4, Supporting information Fig. S6B). As expected, WRCH/DGYW motifs within V segments used by TCR αC strongly correlate with those used by TCR δC (Pearson correlation, r = 0.94; p < 0.001). However, TCR αδV-TCR αC mutated significantly more than other canonical TCR chains (beta: p = 0.0006; gamma: p = 0.007). Mutation in TCR αδV-TCR αC was biased toward C/G mutations (57%) and slightly biased toward transitions (45%) (Table 2). Most C/G mutations occurred within WRCH/DGYW motifs, especially in CDR (FR: 58%, CDR: 71%; 57% overall). TCR αδV-TCR δC mutation also was biased toward C and G nucleotides (63%) within WRCH/DGYW motifs (55%) and toward transition mutations, despite having a much lower rate of mutation (53%; Fig. 3 and 4; Table 2). Replacement mutations occurred significantly more often than silent mutation regardless of constant region utilized (alpha: R/S = 2.2; delta: R/S = 2.48). Generally, TCR αδV gene segments incurred more tandem base mutations than all other chains except perhaps IgHV.
Our previous data suggested that mutation to TCRα may be higher in thymus than in peripheral lymphoid tissues (spleen, spiral valve, and blood), but the limited data set constrained our ability to find a significant difference between tissue types [34]. Thus, we attempted to evaluate any difference in mutation between primary and secondary lymphoid tissues here. We compared frequency of mutation in clones originating from thymus tissue to those originating from peripheral lymphoid tissues. Unfortunately, even our larger dataset constrained analysis. We identified four groups of sequences with identical CDR3 that contained clones from both the thymus and the periphery. Unfortunately, we observed no mutation to any of these sequence groups, so we were unable to compare tissues directly. Using our entire dataset, we analyzed mutation separately for sequences derived from thymus and from peripheral lymphoid tissues. For TCRα, TCRαδ, TCRβ, and TCRγ, results suggested that peripheral lymphoid tissues have a higher frequency of mutation (Supporting information Table S4). However, because we are unable to directly assess mutation in clones derived from common progenitors found in both thymus and peripheral tissues, we hesitate to definitively claim that additional mutation occurs after T cells arrive in the periphery. Further experiments are necessary to ascertain whether mutation frequencies of non-alpha Vs differ between thymus and periphery. Specifically, we need germline genomic Vs and a much deeper CDR3 family clone analysis, but unfortunately no nurse shark genome exists at this time.
Mutation only minimally affects NAR-TCR and IgHV-TCR δC rearrangements
We observed significantly less mutation in both variable domains of NAR-TCR (NTCR V and STCR δV) compared to alpha chain V (NTCR V: p = 0.02; STCR δV: p = 0.01). This lack of mutation confirms earlier reports that NAR-TCR does not utilize SHM [28]. We also found that silent mutation occurred nearly as often as replacement mutation (NTCR V: p = 0.13; STCR δV: p = 0.22; Fig. 3 and 4, Supporting information Fig. S6C). Interestingly, mutations in FR tended to be transversions (NTCR V: 47%; STCR δV: 55%), while those in CDR were biased toward transitions (NTCR V: 63%; STCR δV: 67%; Table 2). Rearrangements between IgHV and TCR constant regions (IgHV-TCR δC) contained more mutation in CDR2 (0.0066) than other regions, and mutation frequency was greater in IgHV-TCR δC than all other TCR V segment types except alpha (TCR αδV-TCR αC; Fig. 3 and 4, Supporting information Fig. S6D). However, mutation in IgHV was substantially lower than would be expected during an antigen-specific response in B cells as they affinity mature in spleen [39, 41, 42]. Mutations to CDR were biased toward G and C nucleotides (71%) within WRCH/DGYW motifs (59%) and tended to be transitions (51%). However, G and C mutation within FR was much lower (49%) and only occurred in motifs 38% of the time (Table 2). Mutations to IgHV-TCR δC typically caused aa replacement at that position (R/S = 3.4, p = 0.02).
Discussion
We previously published the novel use of SHM in TCRα chain of αβ T cells within the thymus of nurse sharks [34]. The frequency of mutation at α was similar to that seen in BCR loci in sharks and mammals. As in B cells, SHM in shark T cells appeared catalyzed by AID and resulted in both point and tandem mutations that accumulated nonconservative aa replacements within antigen-binding regions (CDR) of receptors. However, unlike B cells that use SHM for affinity maturation after exposure to antigen, shark T cells instead use SHM for repertoire diversification during T-cell development within the thymus, implying that SHM contributes to receptor modifications that enhance selection. Here, we extend these findings with an analysis of SHM in other canonical TCR chains, including TCRβ chain of αβ T cells and TCRγ and TCRδ chains of γδ T cells, as well as in noncanonical TCR that associate Ig or Ig-like variable (V) segments with TCR constant (C) regions.
In our previous report, we described an overall mutation frequency of 0.0225 S/N in TCRα chain, with 66% of all mutations to G and C nucleotides [34]. Here, we observed an overall mutation rate of 0.0055 S/N in TCR αδV associated with TCR αC. While these frequencies are considerably lower than those we found before, they reflect the much larger data set used in the current study and likely are more representative of actual substitution rates within TCRα chain. Again, we observed that mutation within TCR αδV-TCR αC affected C and G nucleotides 57% of the time, and 70% of these mutations occurred within CDR and targeted WRCH/DGYW motifs. Mutation was 2.3 times more likely within CDR than framework regions (FR), similar to the 1.87 times we reported before, and mutation was biased toward transitions. Although some V segments accumulated mutation at higher rates than others (i.e., TCR αδV2.4 contained no mutation while TCR αδV5 mutated at a rate of 0.026 S/N), it is clear that AID-catalyzed SHM alters TCR alpha chain in nurse shark.
We then assessed whether SHM is used by the other canonical T-cell chains (beta, gamma, and delta) or by the noncanonical receptor chains that associate IgV segments with TCR C regions (e.g., IgHV-TCR δC rearrangements, doubly rearranging NAR-TCR, and Ig-like TAIL V; see, Fig. 1). To fairly assess mutation patterns between TCRα and TCRδ (since they share a locus), we reclassified and renumbered all new and previously published alpha and delta V segments and identified the constant region associated with each V segment for all sequences within our dataset (see Fig. 2C and Supporting information Fig. S2). We then analyzed mutation data separately for sequences associated with alpha constant regions (TCR αδV-TCR αC) and those associated with delta constant regions (TCR αδV-TCR δC). We found that SHM in nurse sharks significantly alters TCR αδV segments bound to TCR αC but not to TCR δC (p < 0.001). Mutation was found twice as often in TCRα (TCR αδV-TCR αC, 0.0051 S/N) than TCRδ (TCR αδV-TCR δC, 0.0021 S/N) overall, and CDR of TCRα (0.0098 S/N) accumulate more than three times as many mutations as TCRδ CDR (0.0032 S/N). Further, 71% of all G/C mutations to TCR αδV-TCR αC were located within AID-preferred WRCH/DGYW hotspot motifs, compared to 52% in TCR αδV-TCR δC. Mutation in TCR βV and TCR γV reflected similar patterns as TCRδ, with relatively low mutation (TCR βV: 0.0014 S/N; TCR γV: 0.0021 S/N) that is not directed at hotspot motifs (TCR βV: 35%; TCR γV: 30%). The differential use of SHM by TCRα compared to other TCR chains suggests a regulatory control mechanism modulates AID access to the locus during transcription. In mice and humans, V(D)J recombination in Ig heavy chain (IgH: IgM/IgW) V regions is controlled by regulatory elements upstream of the IgH constant (C) region exons (between the V and D clusters) [43, 44]. However, AID accessibility to the IgHC locus is controlled by transcriptional enhancers located in the 3’ regulatory region (RR) downstream of the IgHC exons [43, 45, 46], and SHM requires both the transcription of IgH V regions and the upregulation of AID expression [46]. It is likely that these same transcriptional enhancers regulate SHM of nurse shark TCR, allowing AID access to TCRα chain but limiting access in other TCR chains, evidenced by the greater use of SHM by TCR αδV segments associated with TCRα than with TCRδ constant regions. Since SHM appears to occur in thymus tissue, this differential use of SHM also suggests that AID may be targeting hotspot motifs of TCRα chain to salvage receptors during selection. Roughly half of all beta, gamma, and delta transcripts in our sequence library originated from thymus tissue, and we observed mutation within transcripts from thymus in all TCR chains. Thus, is seems likely that mutation to TCRδ, TCRβ, and TCRγ chain occurs inadvertently as T cells migrate through the thymic cortex during development.
Mutation was low in both V domains — NAR-TCR V and supporting TCR δV — of doubly rearranging NAR-TCR (NTCR V: 0.0023 S/N and STCR δV: 0.0017 S/N). We observed more replacement mutation in NTCR V FR, but replacements did not occur significantly more than silent mutation in either domain (NTCR V: p = 0.13; STCR δV: p = 0.22). Mutation also did not differ between FR and CDR (NTCR V: p = 0.10; STCR δV: p = 0.99) and did not appear to target G and C nucleotides (NTCR V: 41%; STCR δV: 44%) or WRCH/DGYW motifs (NTCR V: 29%; STCR δV: 18%). Mutation in NTCR V and STCR δV mirrors patterns observed in IgH new antigen receptor (IgNAR) transmembrane (Tm) transcripts (0.007 S/N), where mutation has been shown to not target CDR or display bias towards replacements. Though secreted IgNAR forms use SHM for antigen-driven immune responses following antigen stimulation, Tm IgNAR is not used to generate the primary IgNAR repertoire [42, 47]. Diaz et al. suggested the reduced mutation within Tm IgNAR may result from downregulating mutation mechanisms to avoid the risk of creating nonfunctional receptors after four gene rearrangement events [47]. Unusually, mutation in NTCR V and STCR δV favored transversions within FR (NTCR V: 48%; STCR δV: 55%) but transitions within CDR (NTCR V: 63%; STCR δV: 67%). While it is possible that NAR-TCR mutations result unintentionally during thymocyte migration as above, the disparate bias toward transitions and transversions in both domains suggests an alternate or additional process besides AID-catalyzed SHM [48, 49]. Our observations of more limited mutation in TCR γV4 also may indicate that NAR-TCR preferentially pairs with TCR γV4, though no data currently exist to confirm this prediction.
Mutation to V segments of IgHV-TCR δC rearrangements significantly altered nucleotides of CDR2 (p < 0.0001) but not of other regions. Similar to IgHV in B cells, mutation in CDR appeared targeted to G and C nucleotides (71%) within DGWY/WRCH hotspot motifs (59%) and were biased toward transitions (52%). IgHV segments used by TCRs are identical to germline IgHV segments used by B cells, so the similarity in mutation patterns makes sense. Although mutation to IgHV is substantially reduced when associated with a TCR constant region versus a B cell one [39], the significantly greater mutation in CDR2 that is targeted to AID-favored G and C nucleotides suggests that SHM is altering IgHV segments in T cells as well as B cells. The antigen ligands that IgHV-TCR δC receptors bind is not known, nor is the developmental pathway of these cells within the thymus. Nurse shark T cells may simply be using these additional V segments to improve thymic diversity by increasing the pool of Vs available during recombination [34]. Successful rearrangements between an IgHV pseudogene and TCR constant regions support this idea. The motif-rich V domains may be mutated inadvertently as the cells travel through thymic cortical areas where AID is being expressed. However, it also is plausible that these IgHV-T cells are actively modified for simple diversification for free antigen or to salvage receptors during selection that are unable to receive adequate survival signals, though it is unclear whether or not selection is required by this cell type to exit the thymus. Additional studies that assess the anatomical location of developing double-negative thymocytes in addition to functional studies of receptors are necessary to discern further why these V domains are altered.
Recent studies in nonmouse/human organisms confirm the versatility of the TCRαδ locus in the vertebrate immune system. Besides IgHV-TCR δC, TAIL V and NAR-TCR rearrangements in shark, TCRμ of monotremes and marsupials is a hybrid receptor derived from Ig and TCR components [21]. Isoform TCRμ2.0 requires the rearrangement (and in platypus, recombination) of two V domains and is structurally analogous to the doubly rearranging NAR-TCR in nurse sharks [18, 28]. While the C regions of TCRμ resemble traditional TCR δC, the V regions are more similar to IgH V genes, and the TCRμ locus itself reflects the tandem cluster arrangement of Ig rather than the translocon arrangement of TCR [19]. In fact, antibody-like TCR δV segments (VHδ) occur in nearly all gnathostome groups: elasmobranchs (nurse sharks), bony fish (coelacanth), amphibians (Xenopus), birds (chickens), and mammals (platypus, Florida manatee) [20–26]. The passerine zebra finch contains VHδ within a conventional TCR-α/δ locus, while Galliform birds house VHδ gene segments within a second nonsyntenic TCR-δ locus [22]. Thus, the sharing of antigen receptor gene segments seems to be a primordial condition, where the basic immune system started as a “big soup” of receptor parts that, in some lineages, became more and more divergent (and arguably, constrained) as vertebrates moved to land and evolved warm-blooded systems.
Other examples exist to suggest that not only are Ig and TCR gene components interchangeable, but mechanisms of receptor diversification also are shared between B and T cells. In camelids, both gamma- and delta-V gene segments employ SHM to improve structural stability of receptors rather than antigen selection [31]. Analyses included only sequences from peripheral lymphoid tissues, but mutation patterns are parallel to those of TCR γV and TCR αδV-TCR δC in our study. While overall mutation was fairly low (and did not favor G/C nucleotides), replacement mutation was biased toward AID hotspot motifs and resulted in greater length and diversity of CDR3 [31, 32]. Crystalline structures of γδ TCR bound to nonclassical MHC demonstrate that MHC recognition occurs through direct contact by CDR3 of TCR delta [50], suggesting that cells with longer or more diverse CDR3 may be selected for survival. We have evidence that IgHV-TCR δC rearrangements also create longer CDR3 by incorporating one or two diversifying (D) segments from Ig in addition to the single D and J segments from TCR delta (IgV-IgD1-(IgD2)-TCR δD-TCR δJ) during recombination (data not shown). While MHC presentation is not obligatory for γδ T cells, receptors may require stimulation by cell-to-cell contact [51]. Thus, SHM may provide a tool to fine tune receptors to recognize particular antigens, and since both gamma and delta chains are preloaded with numerous hotspot motifs, opportunistic mutation along the V segment is likely.
As we learn more about the immune systems of nonmodel vertebrates, it becomes clearer that ancient lymphocytes likely did not follow the unambiguous rules of B- and T-cell biology found in modern textbooks, and we only just are beginning to understand the myriad schema different vertebrate groups evolved to diversify Ig and TCR repertoires. For example, lymphocyte rearrangement and diversification mechanisms predate the primordial “big bang” of IgSF-based adaptive immunity, as AID-like APOBEC mutators (i.e., CDA1 and CDA2) exist in agnathan vertebrates (hagfish and lamprey) to diversify the variable lymphocyte receptors (VLR) of this more ancient lymphocyte adaptive antigen receptor system [16, 52]. The data presented here offer additional clues to the possible evolutionary relationship between the immune systems of agnathan and jawed vertebrates, suggesting a sustained bipartite use of APOBEC family enzymes to diversify humoral and cellular antigen receptor repertoires, with CDA acting upon variable lymphocyte receptors and AID upon Ig and TCR loci.
Materials and methods
Study animals
TCR sequence data used in this study came from six nurse sharks (G. cirratum) collected off the Florida Keys. We harvested peripheral blood and immune tissues (thymus, spleen, spiral valve) after MS-222 (Argent, Redmond, WA) overdose from five sharks (“Joannie,” “Mary Junior,” “White,” “Grumpy,” “Tom Thumb”) at the University of Maryland’s Center of Marine Biotechnology and one shark (“Florence Nightingale”) at Texas A&M University’s College of Veterinary Medicine. We immediately purified RNA with TRIzol reagent (Life Technologies, Carlsbad, CA). We conducted all research under the Florida Fish and Wildlife Commission Special Activity License SAL-18–2013-SR.
Additionally, we incorporated T-cell sequences from published datasets [25, 26, 28, 35, 53–55] and transcripts from an unpublished multitissue Illumina transcriptome to improve analysis of V segment alignments. We reference published sequences by their GenBank accession number in all relevant Figures.
5’ RACE library generation, cloning, and Sanger sequencing
We used 5 μg total RNA to generate a 5’ RACE (Rapid Amplification of cDNA Ends) library using the GeneRacer Kit (Life Technologies) and a 50:50 mix of oligo-dT and random hexamer primers for cDNA synthesis (Superscript III First Strand Synthesis System, Thermo Fisher Scientific, Inc., Waltham, MA, USA). We estimated cDNA concentration using a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific, Inc.).
We used the GeneRacer 5’ forward primer (Life Technologies) and reverse primers designed to TCR beta (TCRβ), gamma (TCRγ), or delta (TCRδ) constant (C) regions to amplify RACE products for Sanger sequencing (Supporting information Table S4). We followed protocols outlined in [34] for PCR conditions, product visualization, and cloning. We submitted either plasmids or purified PCR products for Sanger sequencing to the DNA Technologies Core Lab on the Texas A&M University campus (College Station, TX, USA) or to GENEWIZ (South Plainfield, NJ, USA). We deposited annotated sequences into the National Center for Biotechnology Information’s (NCBI) GenBank sequence database with the following accession numbers: MN748005-MN748891 and MN788155-MN788287.
Sequence alignment
We used Geneious (version 9.1.8, Biomatters Inc., Auckland, NZ) bioinformatics software to manage DNA sequence data following the same methods as Ott et al. [34]. Sequence alignments and region annotations followed IMGT guidelines [56]. We predicted the location of signal peptide cleavage sites between leader and V segment sequences using SignalP (version 5.0) [57]. A V segment included all bases from the first predicted nucleotide of the V segment to the conserved cysteine (C) residue (YxC motif) at position 104 (“V only”). A V region included all bases between the first predicted nucleotide of the V segment to the last predicted nucleotide of the J segment (V and J). The CDR3 included all bases after the conserved cysteine of the V segment and before the conserved phenylalanine (F) residue (FGxG motif) of the J segment.
We grouped sequences into unique V families based on 70% nucleotide sequence identity and further refined groups into subfamilies based on 80% nucleotide identity [55, 58]. For beta and gamma Vs, we revised the V-segment numbering scheme used by Criscitiello et al. [26] to reflect these parameters. However, we integrated alpha- and delta-V segments into a single group of “αδ Vs” (TCR αδV) to more clearly characterize the locus and avoid confusing name replication within the data. We then numbered the segments according to their position on a phylogenetic tree, from the most divergent branch (TCR αδV1) to the most recent branch (TCR αδV11). For NAR-TCR V segments, we reassigned numbers using this same strategy and renamed the supporting TCR δV segments “STCR δV” to distinguish them from the canonical TCR αδV segments in alignments. We used the same locus-informed numbering system for IgH V identity [53] for all IgHV-TCR δC and IgHV-TCR αC rearrangements. We retained constant region identity (TCR αC/TCR δC) for use in mutation analyses. This system allowed us to clarify V segment usage and analyze differences in mutation patterns for the entire locus. Sequence names followed IMGT unique numbering standards for TCRs [56, 59, 60]. Sequence clone names signify the individual shark (letter following V segment [e.g., TCR γV1X] J = Joannie, M = Mary Junior, G = Grumpy, F = Florence Nightingale, T = Tom Thumb, or W = White] and the tissue type (letter preceding clone number [e.g., T19]: T = thymus, S = spleen, B = peripheral blood leukocytes, and V = spiral valve [intestine]) from which the clone came. For previously published sequences, we use the accession numbers as clone names.
Our preliminary dataset contained 761 TCR αδV, 229 TCR βV, 158 TCR γV, 195 IgMV, 77 IgWV, 11 TAIL V, and 113 NAR-TCR V sequences. For individual Vs within a group (subfamily or allele), we generated a consensus sequence by evaluating sequences found in multiple tissue types and/or sharks, assuming if the exact V nucleotide sequence appeared in more than one tissue or individual that it did not result from the same clone. We equated this consensus sequence to the germline sequence. For all “trans”-rearrangements, we compared mutation in IgHV segments to published germline IgHV sequences [53]. We considered only mutations to IgHV segments associated with TCR δC (not resulting from affinity maturation of B cells). Finally, we compared the incidence of AID hotspot motifs (WRCH/DGYW) between V segments that exhibited mutation and those that lacked mutation to determine the likelihood that AID might catalyze these changes.
Mutation analysis
We analyzed mutation data in beta, gamma, and alpha/delta variable (V) gene segments using methods described by Ott et al. [34] for alpha chain. Mutation frequency was the number of nucleotide changes divided by the total number of nucleotides within a region (e.g., FR, CDR, J, and C), based on differences to a consensus sequence, and recorded disagreements to the consensus as synonymous (S, aa unaltered) or nonsynonymous (N, aa altered). Although insertions and deletions (indels) likely result from similar DNA repair mechanisms as single and tandem mutations, we did not include indels in our mutation data and removed them from nucleotide alignments. However, we indicated the location of each indel in nucleotide alignments by highlighting the nucleotides to either side of the indel (Supporting information Fig. S1–S4). Further, individual sequences that shared less than 70% identity to any group were excluded from mutational analyses. We indicate these sequences using an asterisk next to the clone name within the alignment. We used one-tailed, unpaired Student’s t-test to compare mutation rates between alpha V (TCR αδV-TCR αC) and other V segment types.
To assess whether mutation was AID mediated, we examined V segment consensus sequences for the AID-favored ProSite motifs WRCH/DGYW (G:C mutable target) (International Union of Pure and Applied Chemistry, Zürich, Switzerland; see Sigrist et al. [61]). These motifs serve as common “hotspots” for SHM, where AID favors the G/C bases within WRCH/DGYW motifs [29, 37, 62]. We counted motifs present in consensus sequences (rather than those created by mutation) and compared mutations within clone sequences to target nucleotides within the motif. We defined the frequency of hotspot mutation as the number of mutations to target nucleotides within hotspots for a region (FR or CDR) divided by the total number of mutations in that region and compared FR and CDR regions using χ2 analysis.
We calculated a mutability index for each nucleotide as the observed number of mutations of a specific nucleotide divided by the expected number of mutations of that nucleotide, with a value of 1.00 indicating random mutation [29]. We derived the expected number of mutations by multiplying the frequency of a particular nucleotide within a family of sequences by the total number of observed mutations within that family. We used χ2 analysis to compare mutability indices between FR and CDR regions.
Supplementary Material
Acknowledgments:
We thank the efforts of Kaitlyn Romoser and Ruth Scego for their persistence in sequencing and then annotating hundreds of T cell receptor sequences and the Comparative Immunogenetics Lab at Texas A&M University (Christian Mitchell, Minal Jamsandekar, Brooke Norwood, and Omar Manzur) for their helpful edits. Work was supported by grants NSF IOS 1257829 and IOS-1656870 to MFC.
Abbreviations:
- AID:
activation-induced (cytidine) deaminase
- C:
constant region
- FR:
framework region
- IgH V:
immunoglobulin heavy chain V (IgM/IgW)
- IgNAR:
immunoglobulin heavy chain new (or nurse shark) antigen receptor
- IgSF:
immunoglobulin superfamily
- TAIL V:
TCR associated immunoglobulin-like V
- NTCR V:
NAR T cell receptor distal V domain
- R:
replacement (nonsynonymous) mutation
- S:
silent (synonymous) mutation
- SHM:
somatic hypermutation
- S/N:
substitutions per nucleotide
- STCR δV:
NAR T cell receptor supporting (proximal) δV domain
- TCRδC:
TCR delta constant region
- TCRαC:
TCR alpha constant region
- TCRαδV:
TCR alpha/delta variable segment
- TCRβV:
TCR beta variable segment
- TCRγV:
TCR gamma variable segment
- V:
variable segment
- VHδ:
Ig-like TCR-δ V segments
Footnotes
Conflict of Interest: The authors declare no commercial or financial conflict of interest.
References
- 1.Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT and Litman GW, α, β, γ and δT-cell antigen receptor genes arose early in vertebrate phylogeny. Immunity 1997. 6: 1–11. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein RM, Schluter SF, Lake DF and Marchalonis JJ, Evolutionary conservation and molecular cloning of the recombinase activating gene 1. Biochem. Biophys. Res. Commun. 1994. 205: 687–692. [DOI] [PubMed] [Google Scholar]
- 3.Kasahara M, Vazquez M, Sato K, McKinney EC and Flajnik MF, Evolution of the major histocompatibility complex: isolation of class II A cDNA clones from the cartilaginous fish. PNAS 1992. 89: 6688–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Criscitiello M and Flajnik M, Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur. J. Immunol. 2007. 37: 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flajnik MF, Comparative analyses of immunoglobulin genes: surprises and portents. Nat. Rev. Immunol. 2002. 2: 688–698. [DOI] [PubMed] [Google Scholar]
- 6.Rast JP and Litman GW, T-cell receptor gene homologs are present in the most primitive jawed vertebrates. PNAS 1994. 91: 9248–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schatz DG, V(D)J recombination. Immunol. Rev. 2004. 200: 5–11. [DOI] [PubMed] [Google Scholar]
- 8.Hsu E, Immune system receptors in vertebrates: immunoglobulins. Reference module in life science, Elsevier, Amsterdam, Netherlands, 2018. [Google Scholar]
- 9.Jhunjhunwala S, van Zelm MC, Peak MM and Murre C, Chromatin architecture and the generation of antigen receptor diversity. Cell 2009. 138: 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litman GW, Anderson MK and Rast JP, Evolution of antigen binding receptors. Annu. Rev. Immunol. 1999. 17: 109–147. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D and Scharff MD, The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes. Dev. 2004. 18: 1–11. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani S, Palermo B, Garbelli S, Campanelli R, Robustelli Della Cuna G, Gennari R, Benvenuto F et al. , Dominant TCR-α requirements for a self antigen recognition in humans. J. Immunol. 2002. 169: 6253–6260. [DOI] [PubMed] [Google Scholar]
- 13.Huesmann M, Scott B, Kisielow P and von Boehmer H, Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell 1991. 66: 533–540. [DOI] [PubMed] [Google Scholar]
- 14.Bedel R, Matsuda JL, Brigl M, White J, Kappler J, Marrack P and Gapin L, Lower TCR repertoire diversity in Traj18-deficient mice. Nat. Immunol. 2012. 13: 705–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgulya P, Kishi H, Uematsu Y and von Boehmer H, Exclusion and inclusion of alpha and beta T cell receptor alleles. Cell 1992. 69: 529–537. [DOI] [PubMed] [Google Scholar]
- 16.Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A and Cooper MD, Dual nature of the adaptive immune system in lampreys. Nature 2009. 459: 796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrie HT, Livak F, Schatz DG, Strasser A, Crispe IN and Shortman K, Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J. Exp. Med. 1993. 178: 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra ZE, Baker ML, Schwarz R, Deakin J, Lindblad-Toh K and Miller RD, A unique T cell receptor discovered in marsupials. Proc. Natl. Acad. Sci. USA 2007. 104: 9776–9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Parra ZE and Miller RD, Platypus TCRμ provides insight into the origins and evolution of a uniquely mammalian TCR locus. J. Immunol. 2011. 187: 5246–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breaux B, Hunter ME, Cruz-Schneider MP, Sena L, Bonde RK and Criscitiello MF, The Florida manatee (Trichechus manatus latirostris) T cell receptor loci exhibit V subgroup synteny and chain-specific evolution. Develop. Comparat. Immunol. 2018. 85: 71–85. [DOI] [PubMed] [Google Scholar]
- 21.Parra ZE, Baker ML, Hathaway J, Lopez AM, Trujillo J, Sharp A and Miller RD, Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genomics 2008. 9: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra ZE, Mitchell K, Dalloul RA and Miller RD, A second TCRdelta locus in Galliformes uses antibody-like V domains: insight into the evolution of TCRdelta and TCRμ genes in tetrapods. J. Immunol. 2012. 188: 3912–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF and Miller RD, The dynamic TCRdelta: TCRdelta chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur. J. Immunol. 2010. 40: 2319–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, Buonocore F et al. , Genome complexity in the coelacanth is reflected in its adaptive immune system. J. Experiment. Zool. Part B Mol. Develop. Evol. 2014. 322: 438–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deiss TC, Breaux B, Ott JA, Daniel RA, Chen PL, Castro CD, Ohta Y et al. , Ancient use of Ig variable domains contributes significantly to the TCRδ repertoire. J. Immunol. 2019. 203: 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Criscitiello MF, Ohta Y, Saltis M, McKinney EC and Flajnik MF, Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J. Immunol. 2010. 184: 6950–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y et al. , Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014. 505: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Criscitiello M, Saltis M and Flajnik M, An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. PNAS 2006. 103: 5036–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Bernstein H, Ranganathan P and Schluter S, Somatic hypermutation of TCR γ V genes in the sandbar shark. Dev. Comp. Immunol. 2012. 37: 176–183. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Kshirsagar S, Jensen I, Lau K, Covarrubias R, Schluter SF and Marchalonis JJ, Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc. Natl. Acad. Sci. USA 2009. 106: 8591–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciccarese S, Vaccarelli G, Lefranc MP, Tasco G, Consiglio A, Casadio R, Linguiti G and Antonacci R, Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma (TRG) and delta (TRD) variable domains. Dev. Comp. Immunol. 2014. 46: 300–313. [DOI] [PubMed] [Google Scholar]
- 32.Antonacci R, Mineccia M, Lefranc MP, Ashmaoui HM, Lanave C, Piccinni B, Pesole G et al. , Expression and genomic analyses of Camelus dromedarius T cell receptor delta (TRD) genes reveal a variable domain repertoire enlargement due to CDR3 diversification and somatic mutation. Mol. Immunol. 2011. 48: 1384–1396. [DOI] [PubMed] [Google Scholar]
- 33.Vaccarelli G, Antonacci R, Tasco G, Yang F, Giordano L, El Ashmaoui HM., Hassanane MS. et al. , Generation of diversity by somatic mutation in the Camelus dromedarius T-cell receptor gamma variable domains. Eur. J. Immunol. 2012. 42: 3416–3428. [DOI] [PubMed] [Google Scholar]
- 34.Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF and Criscitiello MF, Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus. eLife 2018. 7: e28477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malecek K, Lee V, Feng W, Huang JL, Flajnik MF, Ohta Y and Hsu E, Immunoglobulin heavy chain exclusion in the shark. PLoS Biol 2008. 6: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flajnik MF, Deschacht N and Muyldermans S, A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011. 9: e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogozin IB and Diaz M, Cutting Edge: DGYW/WRCH is a better predictor of mutability at G:C Bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 2004. 172: 3382–3384. [DOI] [PubMed] [Google Scholar]
- 38.Anderson MK, Shamblott MJ, Litman RT and Litman GW, Generation of immunoglobulin light chain gene diversity in Raja erinacea is not associated with somatic rearrangement, an exception to a central paradigm of B cell immunity. J. Exp. Med. 1995. 182: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SS, Tranchina D, Ohta Y, Flajnik MF and Hsu E, Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 2002. 16: 571–582. [DOI] [PubMed] [Google Scholar]
- 40.Rumfelt L, McKinney E, Taylor E and Flajnik M, The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol. 2002. 56: 130–148. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Du Pasquier L and Hsu E, Shark IgW C region diversification through RNA processing and isotype switching. J. Immunol. 2013. 191: 3410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dooley H, Stanfield RL, Brady RA and Flajnik MF, First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. PNAS 2006. 103: 1846–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B et al. , The IgH 3’ regulatory region controls somatic hypermutation in germinal center B cells. J. Exp. Med. 2013. 210: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng H-L, Hansen E et al. , CTCF-binding elements mediate control of V(D)J recombination. Nature 2011. 477: 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P and Papavasiliou FN, Switch recombination and somatic hypermutation are controlled by the heavy chain 3 enhancer region. J. Exp. Med. 2009. 206: 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Komori A, Xu Z, Wu X, Zan H and Casali P, Biased dA/dT somatic hypermutation as regulated by the heavy chain intronic iEmu enhancer and 3’Ealpha enhancers in human lymphoblastoid B cells. Mol. Immunol. 2006. 43: 1817–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz M, Greenberg A and Flajnik M, Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. PNAS 1998. 95: 14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krijger PH, Tsaalbi-Shtylik A, Wit N, van den Berk PCM, de Wind N and Jacobs H, Rev1 is essential in generating G to C transversions downstream of the Ung2 pathway but not the Msh2+Ung2 hybrid pathway. Eur. J. Immunol. 2013. 43: 2765–2770. [DOI] [PubMed] [Google Scholar]
- 49.Thientosapol ES, Bosnjak D, Durack T, Stevanovski I, van Geldermalsen M, Holst J, Jahan Z et al. , SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc. Natl. Acad. Sci. 2018. 115: 4921–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams EJ, Chien YH and Garcia KC, Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science 2005. 308: 227–231. [DOI] [PubMed] [Google Scholar]
- 51.Allison TJ and Garboczi DN, Structure of gammadelta T cell receptors and their recognition of non-peptide antigens. Mol. Immunol. 2002. 38: 1051–1061. [DOI] [PubMed] [Google Scholar]
- 52.Alder MN, Herrin BR, Sadlonova A, Stockard CR, Grizzle WE, Gartland LA, Gartland GL et al. , Antibody responses of variable lymphocyte receptors in the lamprey. Nat. Immunol. 2008. 9: 319–327. [DOI] [PubMed] [Google Scholar]
- 53.Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A and Hsu E, The evolution of multiple isotypic IgM heavy chain genes in the shark. J. Immunol. 2008. 180: 7461–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF and Hsu E, Somatic hypermutation and junctional diversification at Ig heavy chain loci in the nurse shark. J. Immunol. 2005. 175: 8105–8115. [DOI] [PubMed] [Google Scholar]
- 55.Rumfelt LL, Lohr RL, Dooley H and Flajnik MF, Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004. 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V and Lefranc G, IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003. 27: 55–77. [DOI] [PubMed] [Google Scholar]
- 57.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G and Nielsen H, SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019. 37: 420–423. [DOI] [PubMed] [Google Scholar]
- 58.Brodeur P and Riblet R, The immunoglobulin heavy chain variable region (Igh-V) locus in the mouse. I. One hundred Igh-V genes comprise seven families of homologous genes. Eur. J. Immunol. 1984. 14: 922–930. [DOI] [PubMed] [Google Scholar]
- 59.Lefranc M-P, Immunoglobulin and T cell receptor genes: IMGT and the birth and rise of immunoinformatics. Front. Immunol. 2014. 5: 22–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohlin M, Scheepers C, Corcoran M, Lees WD, Busse CE, Bagnara D, Thörnqvist L et al. , Inferred allelic variants of immunoglobulin receptor genes: a system for their evaluation, documentation, and naming. Front. Immunol. 2019. 10: 435–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A and Hulo N, PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010. 38: D161–D166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei L, Chahwan R, Wang S, Wang X, Pham PT, Goodman MF, Bergman A et al. , Overlapping hotspots in CDRs are critical sites for V region diversification. Proc. Natl. Acad. Sci. 2015. 112: E728–E737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.