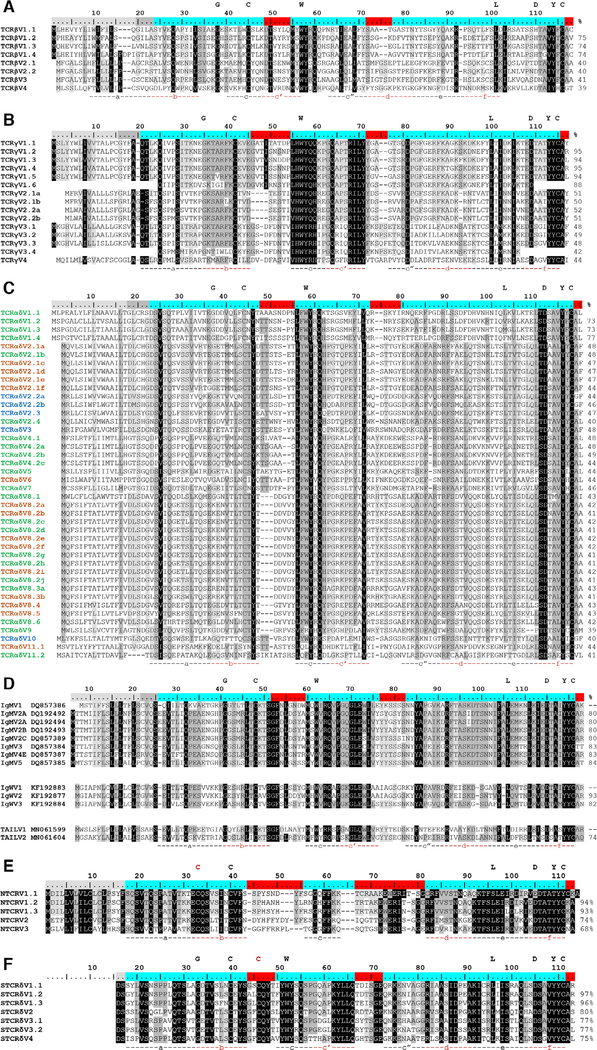
Figure 2.
Consensus sequence alignments for T-cell receptor V segments indicate substantial conservation between segments. V gene segments are grouped by identity for (A) beta (TCRβV) (B) gamma (TCRγV), (C) alpha/delta (TCRαδV), (D) immunoglobulin (Ig, IgMV and IgWV) and TCR-associated Ig-like V (TAILV), (E) NAR-TCR distal V domain (NTCRV), and (F) NAR-TCR proximal Vδ domain (STCRδV). V segment families share >70% nucleotide identity (e.g., TCRαδV2) and subfamilies have >80% nucleotide identity (e.g., TCRαδV2.1). Alleles share >90% nucleotide identity and common differences appear in more than one shark (e.g., TCRαδV2.1a). Letters above the scale denote conserved residues of antigen receptor domains. Regions below the alignment designate predicted beta strand location and direction. Shading within an alignment indicates amino acid conservation (Blosum62 score matrix [Threshold = 1]: black = 100% similar; dark grey = 80–100%; light grey = 60–80%). Values to the right of the alignments show the percent nucleotide identity to the first sequence. Highlighting within the scale indicates leader peptides (gray), framework regions (blue), and complementarity-determining regions (CDR, red). Coloring within the TCRαδV consensus sequence names identify the constant region used (green = TCR αC; blue = TCR δC; orange = both TCR αV and TCR δC). IgM or IgW germline sequence accession numbers are in sequence titles. IgM V2C is an Ig pseudogene due to defective Ig constant region exons but can form functional transcripts when associated with TCRα or TCRδ C. Each NAR-TCR V domain is encoded by V gene segments from a single gene family, but we employed original names to indicate the NARTCR cluster used. Gaps within a sequence are for alignment purposes only. Data from a single experiment, where each PCR tube represents a single replicate for each chain, shark, and tissue sample (see Supporting information Table 1 and 2).