Fig. 4. CMP can predict relatively accurate experimental binding affinities, across multiple ligands and proteins.
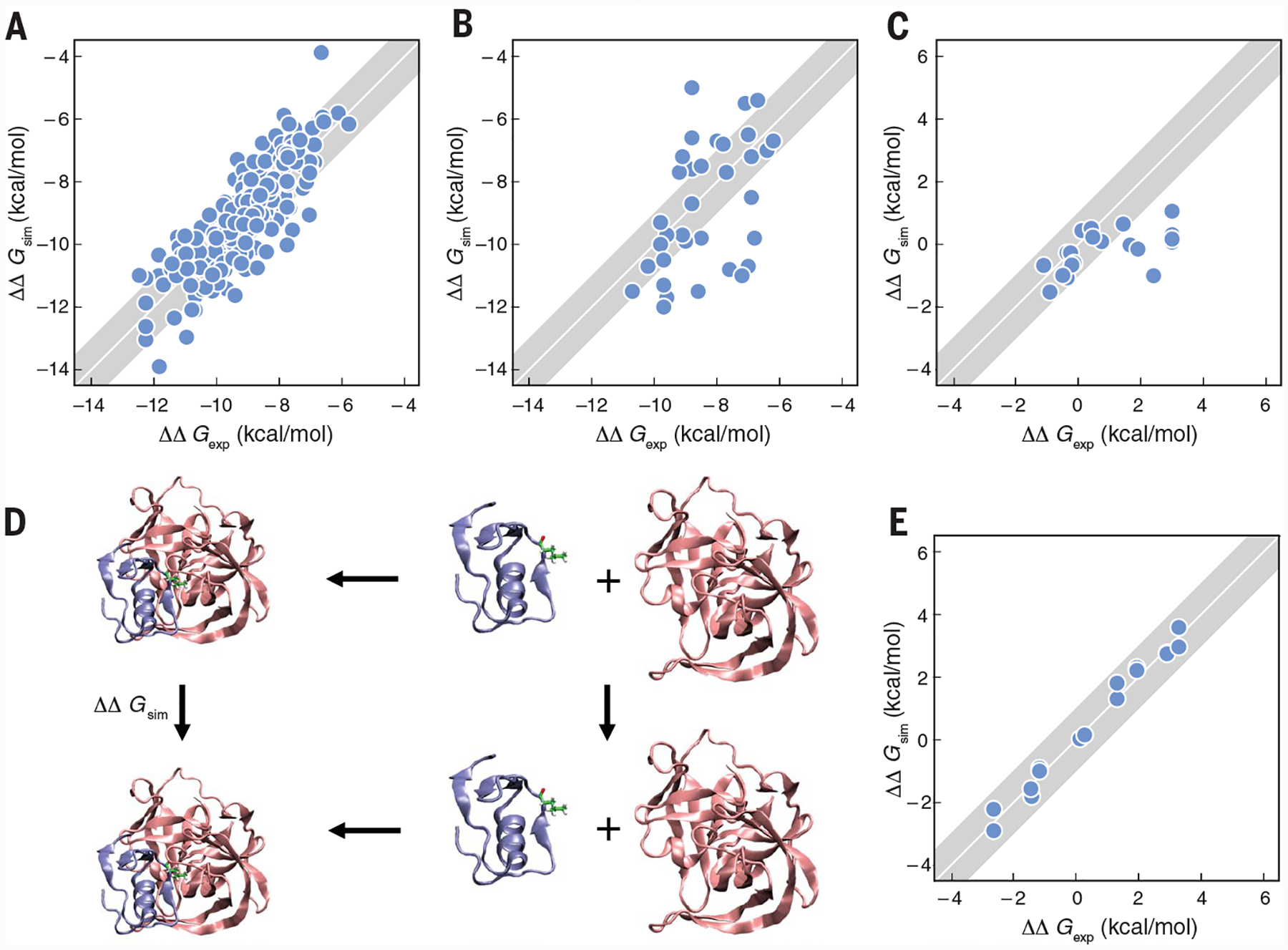
The diagonal line represents perfect agreement with experiments. The shaded area indicate 1 kcal-mol−1 error bars. (A) Schrodinger free-energy perturbation calculations of 200 ligands in eight proteins (108). (B) Algedhi et al. prediction of binding affinities of ligands across protein families (111). (C) MELD × MD relative binding affinities of various P53 mutant peptides, which are highly flexible, to the MDM2/x protein (57). (D) The thermodynamic cycle of Zou et al. for computing mutational effects on protein-protein binding (112). (E) Predicted affinities versus experiments for (D) (112).
