Fig. 1. DNA unwrapping regulates Cas12a cleavage of nucleosomal targets.
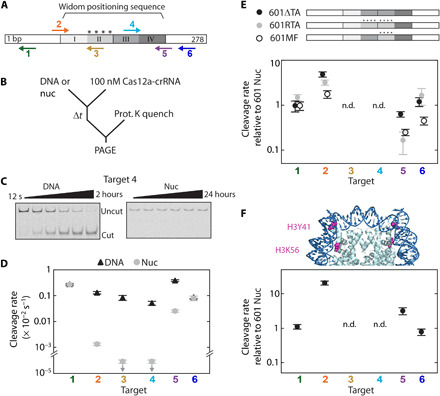
(A) The Widom 601 positioning sequence is divided into quartiles indicating the inner (II and III) and outer (I and IV) wrap and is flanked by DNA. “****”: four TA dinucleotide repeats that produce tight wrapping of the inner left quartile. Arrows: Cas12a targets, pointing in the direction of R-loop formation; for top arrows, R-loop forms with the Crick (bottom) strand, and for bottom arrows, R-loop forms with the Watson (top) strand. (B) Cleavage reaction setup. nuc, nucleosome substrate. (C) Representative gels of target 4 cleavage by Cas12a. (D) Cleavage rates for the six DNA and nucleosome Cas12a targets at 25°C. Downward arrows signify that the value is an upper limit due to the lack of detectable cleavage. (E) Top: Diagram depicting variant 601 constructs (fig. S1E). Bottom: Cas12a cleavage of variant nucleosome substrates normalized to original 601 nucleosome substrate. (F) Top: Crystal structure of the 601 nucleosome [Protein Data Bank (PDB): 3LZ0 (80)] highlighting the amino acid modifications H3Y41E and H3K56Q, which mimic H3Y41ph and H3K56ac. Bottom: Cleavage rates of H3 mutant nucleosome normalized to the original wt nucleosome. n.d., no data, as no cleavage was observed for targets 3 and 4 for all nucleosome substrates. (D to F) Each data point is the mean of at least three replicates; error bars: SEM.
