Fig. 4. Phase-separated nucleosome arrays minimally inhibit DNA accessibility.
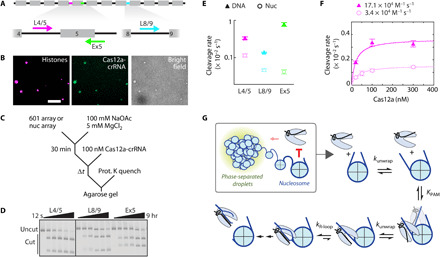
(A) Diagram of a 12-mer 601 array. Gray boxes: 601 positioning sequences; black lines: unique linker DNAs. Cas12a targets are highlighted (L4/5, magenta; Ex5, green; L8/9, cyan), and the associated arrows represent direction of R-loop formation. (B) Phase-separated nucleosome array droplets colocalize with dCas12a-L4/5. Histone H2B is labeled with AF594; L4/5 crRNA is labeled with AF488. Scale bar, 10 μm. (C) Reaction scheme depicting Cas12a cleavage of DNA arrays or phase-separated nucleosome arrays. (D) Representative agarose gel showing Cas12a cleavage of phase-separated nucleosome arrays, detected by ethidium bromide. All reactions are from the same gel; the black line represents cropping of the gel. (E) Cas12a cleavage plot of DNA and nucleosome arrays. Data points are color-coordinated with (A). Rates are included in table S5. (F) Concentration-dependent Cas12a cleavage of L4/5, targeting DNA, and nucleosome arrays. (E and F) Each data point represents the mean of at least three replicates (exception: DNA cleavage at 30 and 200 nM Cas12a was done in duplicate). Error bars: SEM. (G) Model of Cas12a cleavage inhibition by the nucleosome and chromatin. Chromatin has a small impact on Cas12a cleavage efficiency, which can be attributed to neighboring nucleosomes. The nucleosome inhibits Cas12a binding to wrapped DNA.
