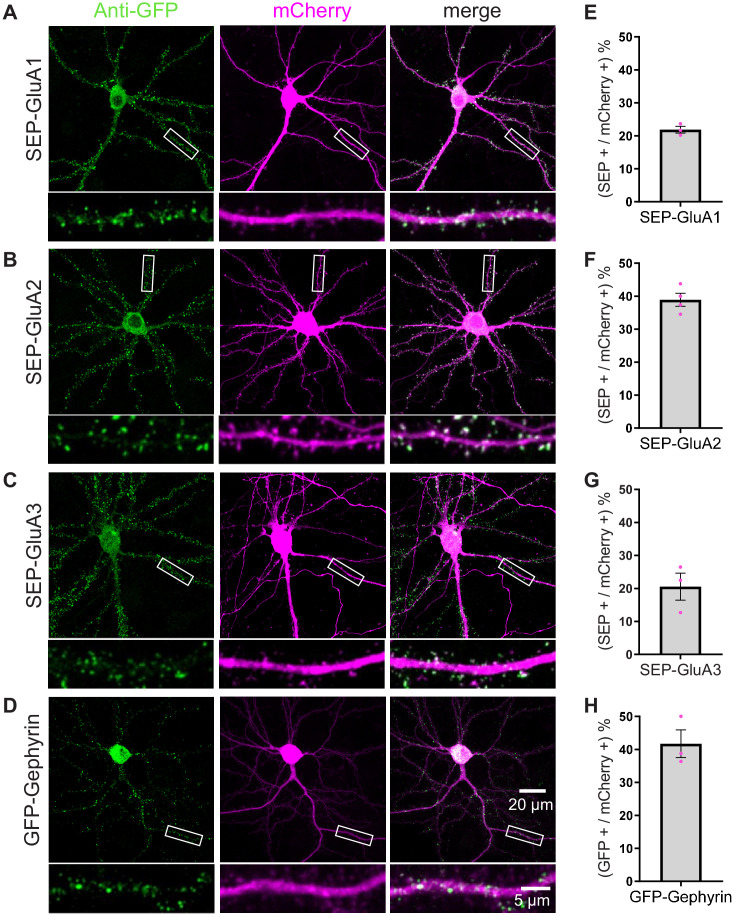
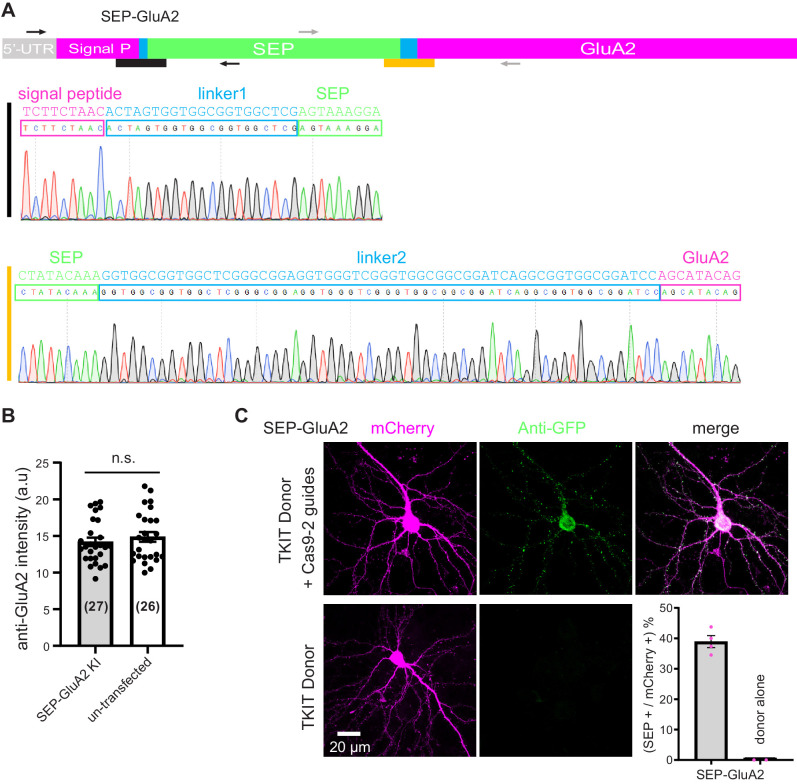
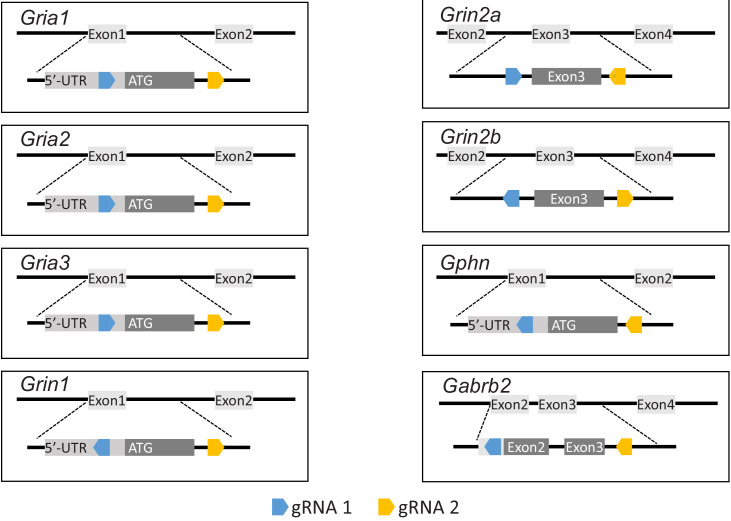
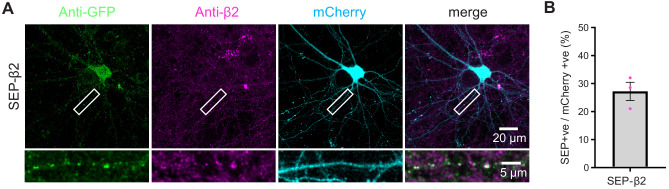
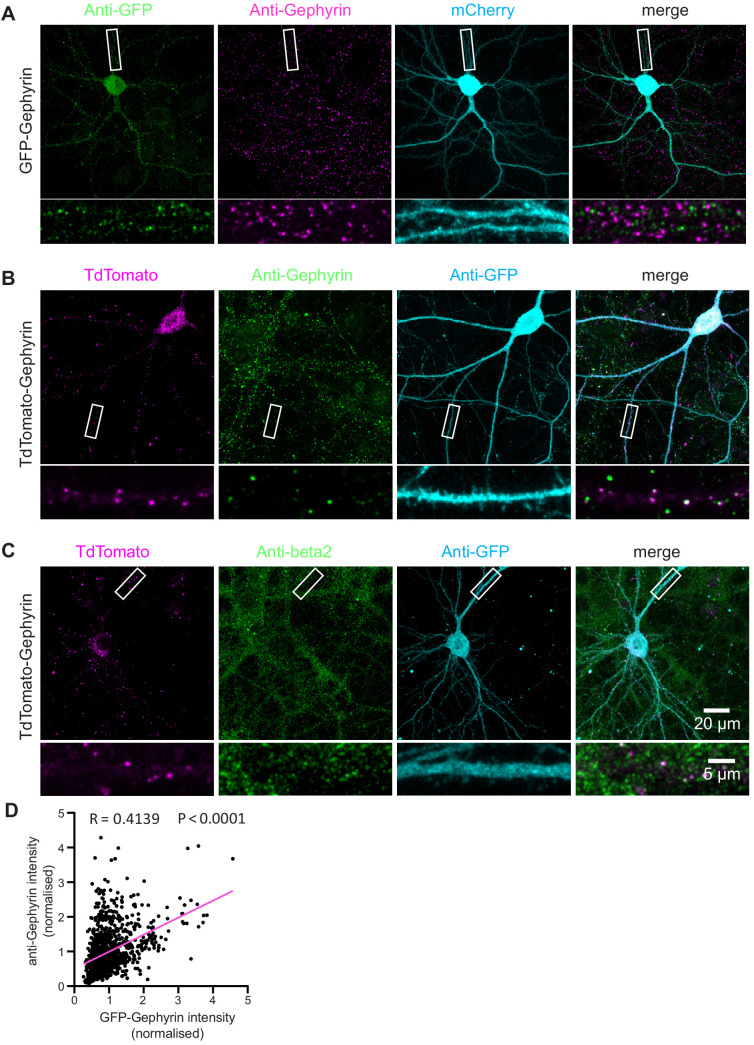
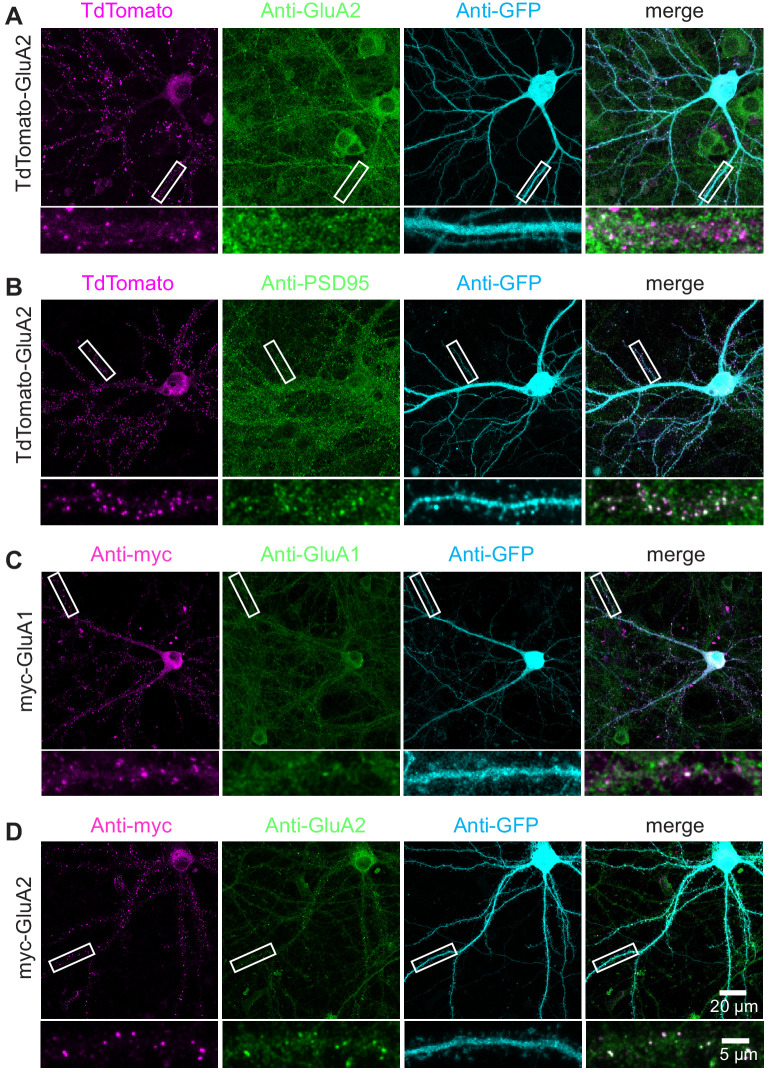
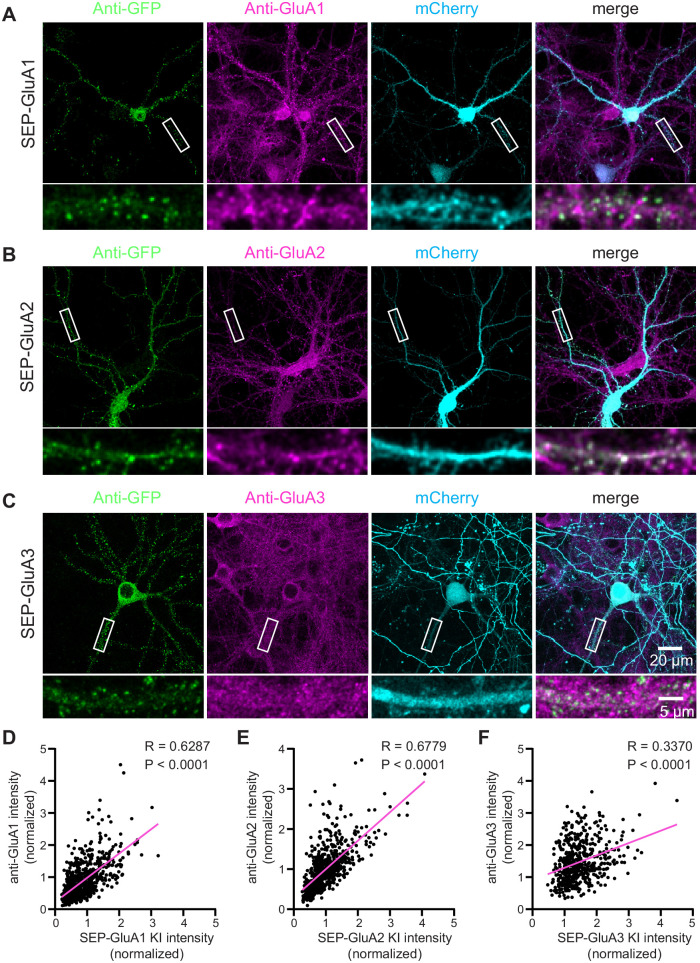
Figure 2. Efficient KI of SEP-GluA1-3 and GFP-gephyrin using TKIT.
(A) SEP-GluA1, (B) SEP-GluA2, (C) SEP-GluA3, and (D) GFP-gephyrin visualized with confocal microscopy after neurons were co-transfected with TKIT constructs and a mCherry cell fill. Segments of dendrite in the white boxed region are enlarged below. Scale bar, 20 μm or 5 μm. Average efficiency of KI for (E) SEP-GluA1, (F) SEP-GluA2, (G) SEP-GluA3, and (H) GFP-gephyrin. Error bars display SEM.