Fig. 6.
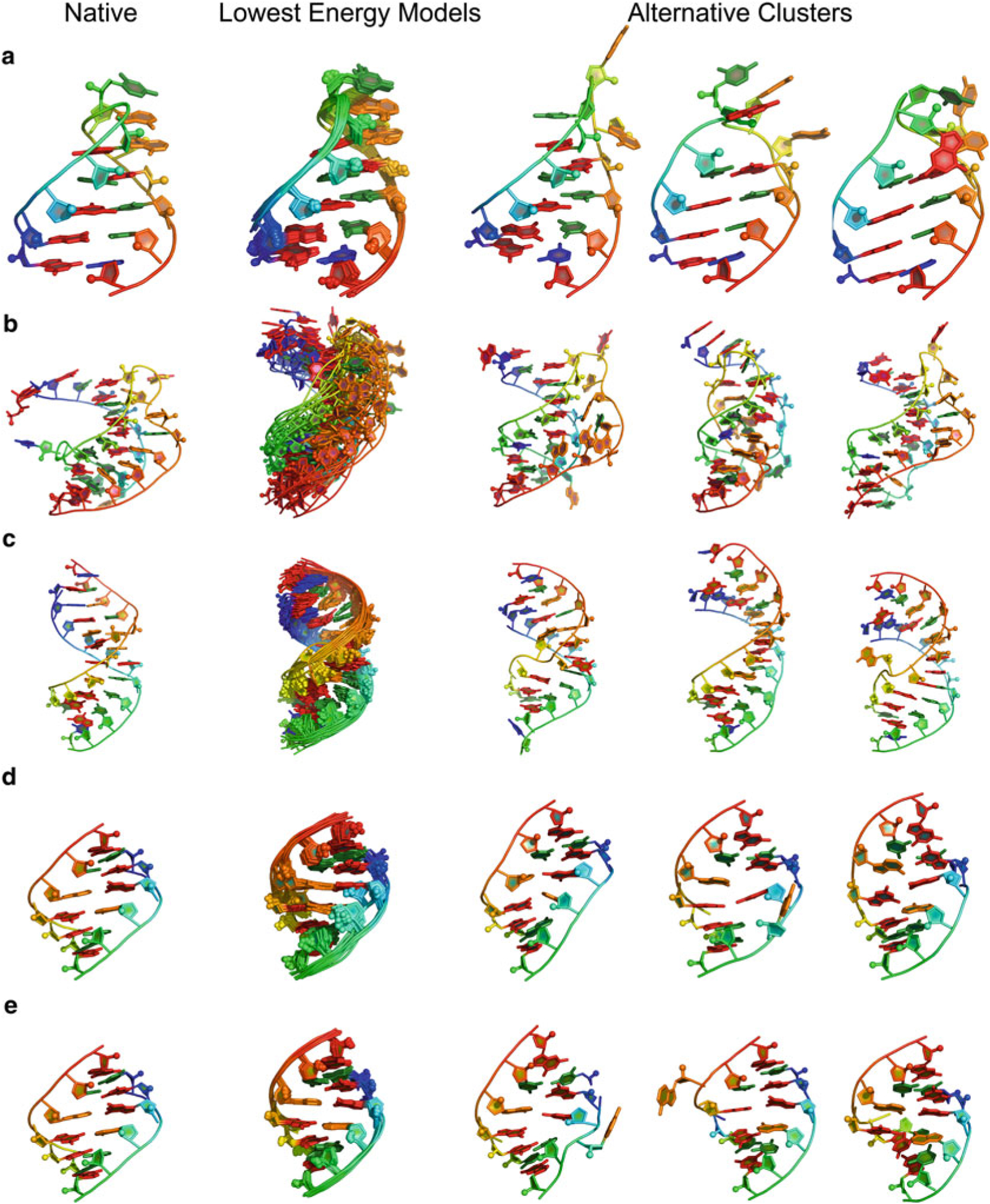
(a) GCAA tetraloop (1ZIH): RNA De Novo (through fragment assembly of RNA with full atom refinement, FARFAR) gives lowest energy models displaying structural convergence. (b) Pseudoknot (1L2X) [27], less converged then tetraloop–but also a larger RNA–gives models that are still within 3 Å heavy-atom RMSD for top model. (c) 4 × 4 internal loop solved by NMR at PDB ID 2L8F [28], converges despite presenting four noncanonical base pairs. (d) Tandem GA (1MIS) [26] without application of 1H chemical shifts. (e) Tandem GA with 1H chemical shifts, demonstrates the improved convergence with the addition of 1H chemical shift
